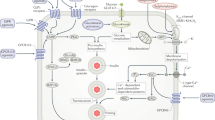
Key Points
-
The list of known variants affecting type 2 diabetes mellitus (T2DM) risk confirms that this disease has a multifactorial aetiology
-
The concept of precision medicine has been exemplified in pharmacogenetic studies of monogenic diabetes mellitus
-
The genetic architecture of mild adverse drug reactions and treatment efficacy for antidiabetic agents probably resembles that of T2DM and other complex traits
-
Existing pharmacogenetic evidence of T2DM is limited; future pharmacogenomic studies utilizing large samples sizes will help identify variants that reveal novel mechanisms of drug action
-
Genetic evidence-based 'dose-response' curves have been used in validating candidate drug targets
-
Pharmacogenomic studies adopting a systems biology approach are expected to provide context specific evidence for future T2DM drug development
Abstract
Genomic studies have greatly advanced our understanding of the multifactorial aetiology of type 2 diabetes mellitus (T2DM) as well as the multiple subtypes of monogenic diabetes mellitus. In this Review, we discuss the existing pharmacogenetic evidence in both monogenic diabetes mellitus and T2DM. We highlight mechanistic insights from the study of adverse effects and the efficacy of antidiabetic drugs. The identification of extreme sulfonylurea sensitivity in patients with diabetes mellitus owing to heterozygous mutations in HNF1A represents a clear example of how pharmacogenetics can direct patient care. However, pharmacogenomic studies of response to antidiabetic drugs in T2DM has yet to be translated into clinical practice, although some moderate genetic effects have now been described that merit follow-up in trials in which patients are selected according to genotype. We also discuss how future pharmacogenomic findings could provide insights into treatment response in diabetes mellitus that, in addition to other areas of human genetics, facilitates drug discovery and drug development for T2DM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Daly, A. K. Pharmacogenomics of adverse drug reactions. Genome Med. 5, 5 (2013).
Zhou, K. & Pearson, E. R. Insights from genome-wide association studies of drug response. Annu. Rev. Pharmacol. Toxicol. 53, 299–310 (2013).
American Diabetes, A. Diagnosis and classification of diabetes mellitus. Diabetes Care 27, S5–S10 (2004).
Stumvoll, M., Goldstein, B. J. & van Haeften, T. W. Type 2 diabetes: principles of pathogenesis and therapy. Lancet 365, 1333–1346 (2005).
Morris, A. P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Prasad, R. B. & Groop, L. Genetics of type 2 diabetes — pitfalls and possibilities. Genes (Basel) 6, 87–123 (2015).
Steinthorsdottir, V. et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat. Genet. 46, 294–298 (2014).
Flannick, J. et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 46, 357–363 (2014).
Mohlke, K. L. & Boehnke, M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum. Mol. Genet. 24, R85–92 (2015).
Holman, R. R., Paul, S. K., Bethel, M. A., Matthews, D. R. & Neil, H. A. 10-year follow-up of intensive glucose control in type 2 diabetes. N. Engl. J. Med. 359, 1577–1589 (2008).
Stratton, I. M. et al. Association of glycaemia with macrovascular and microvascular complications of type 2 diabetes (UKPDS 35): prospective observational study. BMJ 321, 405–412 (2000).
[No authors listed.] Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet 352, 837–853 (1998).
Bailey, C. J. The current drug treatment landscape for diabetes and perspectives for the future. Clin. Pharmacol. Ther. 98, 170–184 (2015).
Deacon, C. F. Dipeptidyl peptidase-4 inhibitors in the treatment of type 2 diabetes: a comparative review. Diabetes Obes. Metab. 13, 7–18 (2011).
Tahrani, A. A., Barnett, A. H. & Bailey, C. J. SGLT inhibitors in management of diabetes. Lancet Diabetes Endocrinol. 1, 140–151 (2013).
Pearson, E. R. et al. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 362, 1275–1281 (2003).
Gloyn, A. L. et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med. 350, 1838–1849 (2004).
Carr, D. F., Alfirevic, A. & Pirmohamed, M. Pharmacogenomics: current state-of-the-art. Genes (Basel) 5, 430–443 (2014).
Goldstein, D. B., Tate, S. K. & Sisodiya, S. M. Pharmacogenetics goes genomic. Nat. Rev. Genet. 4, 937–947 (2003).
Goldstein, D. B. et al. Sequencing studies in human genetics: design and interpretation. Nat. Rev. Genet. 14, 460–470 (2013).
Mizzi, C. et al. Personalized pharmacogenomics profiling using whole-genome sequencing. Pharmacogenomics 15, 1223–1234 (2014).
Zhou, K. et al. Heritability of variation in glycaemic response to metformin: a genome-wide complex trait analysis. Lancet Diabetes Endocrinol. 2, 481–487 (2014).
Yang, J. et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43, 519–525 (2011).
Visscher, P. M. et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 10, e1004269 (2014).
Yang, J. et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 47, 1114–1120 (2015).
Elks, C. E. et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front. Endocrinol. (Lausanne) 3, 29 (2012).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
McCreight, L. J., Bailey, C. J. & Pearson, E. R. Metformin and the gastrointestinal tract. Diabetologia 59, 426–435 (2016).
Simonis-Bik, A. M. et al. Genetic influences on the insulin response of the beta cell to different secretagogues. Diabetologia 52, 2570–2577 (2009).
Gjesing, A. P. et al. High heritability and genetic correlation of intravenous glucose- and tolbutamide-induced insulin secretion among non-diabetic family members of type 2 diabetic patients. Diabetologia 57, 1173–1181 (2014).
De Silva, N. M. & Frayling, T. M. Novel biological insights emerging from genetic studies of type 2 diabetes and related metabolic traits. Curr. Opin. Lipidol. 21, 44–50 (2010).
Daly, A. K. et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 41, 816–819 (2009).
Harper, A. R. & Topol, E. J. Pharmacogenomics in clinical practice and drug development. Nat. Biotechnol. 30, 1117–1124 (2012).
Maranville, J. C. & Cox, N. J. Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharmacogenomics J. http://dx.doi.org/10.1038/tpj.2015.47 (2015).
Hellerstein, M. K. Exploiting complexity and the robustness of network architecture for drug discovery. J. Pharmacol. Exp. Ther. 325, 1–9 (2008).
Nelson, M. R. et al. Genome-wide approaches to identify pharmacogenetic contributions to adverse drug reactions. Pharmacogenomics J. 9, 23–33 (2009).
Zhou, K. et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet. 43, 117–120 (2011).
Becker, M. L., Pearson, E. R. & Tkac, I. Pharmacogenetics of oral antidiabetic drugs. Int. J. Endocrinol. 2013, 686315 (2013).
Semiz, S., Dujic, T. & Causevic, A. Pharmacogenetics and personalized treatment of type 2 diabetes. Biochem. Med. (Zagreb) 23, 154–171 (2013).
Todd, J. N. & Florez, J. C. An update on the pharmacogenomics of metformin: progress, problems and potential. Pharmacogenomics 15, 529–539 (2014).
Shu, Y. et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther. 83, 273–280 (2008).
Graham, G. G. et al. Clinical pharmacokinetics of metformin. Clin. Pharmacokinet. 50, 81–98 (2011).
Zhou, K. et al. Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes 58, 1434–1439 (2009).
Jablonski, K. A. et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes 59, 2672–2681 (2010).
Becker, M. L. et al. Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J. 9, 242–247 (2009).
Shu, Y. et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Invest. 117, 1422–1431 (2007).
Zhou, K. et al. Loss-of-function CYP2C9 variants improve therapeutic response to sulfonylureas in type 2 diabetes: a Go-DARTS study. Clin. Pharmacol. Ther. 87, 52–56 (2010).
Becker, M. L. et al. Cytochrome P450 2C9 *2 and *3 polymorphisms and the dose and effect of sulfonylurea in type II diabetes mellitus. Clin. Pharmacol. Ther. 83, 288–292 (2008).
Suzuki, K. et al. Effect of CYP2C9 genetic polymorphisms on the efficacy and pharmacokinetics of glimepiride in subjects with type 2 diabetes. Diabetes Res. Clin. Pract. 72, 148–154 (2006).
Pearson, E. R. et al. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes 56, 2178–2182 (2007).
Schroner, Z. et al. Effect of sulphonylurea treatment on glycaemic control is related to TCF7L2 genotype in patients with type 2 diabetes. Diabetes Obes. Metab. 13, 89–91 (2011).
Javorsky, M. et al. Association between TCF7L2 genotype and glycemic control in diabetic patients treated with gliclazide. Int. J. Endocrinol. 2013, 374858 (2013).
Javorsky, M. et al. KCNJ11 gene E23K variant and therapeutic response to sulfonylureas. Eur. J. Intern. Med. 23, 245–249 (2012).
Feng, Y. et al. Ser1369Ala variant in sulfonylurea receptor gene ABCC8 is associated with antidiabetic efficacy of gliclazide in Chinese type 2 diabetic patients. Diabetes Care 31, 1939–1944 (2008).
Zhang, H., Liu, X., Kuang, H., Yi, R. & Xing, H. Association of sulfonylurea receptor 1 genotype with therapeutic response to gliclazide in type 2 diabetes. Diabetes Res. Clin. Pract. 77, 58–61 (2007).
Pei, Q. et al. PPAR-γ2 and PTPRD gene polymorphisms influence type 2 diabetes patients' response to pioglitazone in China. Acta Pharmacol. Sin. 34, 255–261 (2013).
Kang, E. S. et al. Effects of Pro12Ala polymorphism of peroxisome proliferator-activated receptor γ2 gene on rosiglitazone response in type 2 diabetes. Clin. Pharmacol. Ther. 78, 202–208 (2005).
Hsieh, M. C. et al. Common polymorphisms of the peroxisome proliferator-activated receptor-γ (Pro12Ala) and peroxisome proliferator-activated receptor-γ coactivator-1 (Gly482Ser) and the response to pioglitazone in Chinese patients with type 2 diabetes mellitus. Metabolism 59, 1139–1144 (2010).
t Hart, L. M. et al. The CTRB1/2 locus affects diabetes susceptibility and treatment via the incretin pathway. Diabetes 62, 3275–3281 (2013).
van Leeuwen, N. et al. A gene variant near ATM is significantly associated with metformin treatment response in type 2 diabetes: a replication and meta-analysis of five cohorts. Diabetologia 55, 1971–1977 (2012).
Zhou, Y. et al. RS11212617 is associated with metformin treatment response in type 2 diabetes in Shanghai local Chinese population. Int. J. Clin. Pract. 68, 1462–1466 (2014).
Connelly, P. J. et al. Recessive mutations in the cancer gene Ataxia Telangiectasia Mutated (ATM), at a locus previously associated with metformin response, cause dysglycaemia and insulin resistance. Diabet. Med. 33, 371–375 (2016).
Gokalp, O. et al. Mild hypoglycaemic attacks induced by sulphonylureas related to CYP2C9, CYP2C19 and CYP2C8 polymorphisms in routine clinical setting. Eur. J. Clin. Pharmacol. 67, 1223–1229 (2011).
Zhang, Y. et al. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on pharmacokinetics of gliclazide MR in Chinese subjects. Br. J. Clin. Pharmacol. 64, 67–74 (2007).
Bailey, S. D. et al. Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study. Diabetes Care 33, 2250–2253 (2010).
Watanabe, I. et al. A study to survey susceptible genetic factors responsible for troglitazone-associated hepatotoxicity in Japanese patients with type 2 diabetes mellitus. Clin. Pharmacol. Ther. 73, 435–455 (2003).
Dujic, T. et al. Association of organic cation transporter 1 with intolerance to metformin in type 2 diabetes: a GoDARTS study. Diabetes 64, 1786–1793 (2015).
Bailey, C. J. Safety of antidiabetes medications: an update. Clin. Pharmacol. Ther. 98, 185–195 (2015).
Kumashiro, R. et al. Association of troglitazone-induced liver injury with mutation of the cytochrome P450 2C19 gene. Hepatol. Res. 26, 337–342 (2003).
Tarasova, L. et al. Association of genetic variation in the organic cation transporters OCT1, OCT2 and multidrug and toxin extrusion 1 transporter protein genes with the gastrointestinal side effects and lower BMI in metformin-treated type 2 diabetes patients. Pharmacogenet. Genomics 22, 659–666 (2012).
Dujic, T. et al. Organic cation transporter 1 variants and gastrointestinal side effects of metformin in patients with type 2 diabetes. Diabet. Med. 33, 511–514 (2016).
Hay, M., Thomas, D. W., Craighead, J. L., Economides, C. & Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 32, 40–51 (2014).
Nelson, M. R. et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013).
Cohen, J. C. Emerging LDL therapies: using human genetics to discover new therapeutic targets for plasma lipids. J. Clin. Lipidol. 7, S1–5 (2013).
Abifadel, M. et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156 (2003).
Cohen, J. C., Boerwinkle, E., Mosley, T. H. Jr & Hobbs, H. H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354, 1264–1272 (2006).
Global Lipids Genetics, C. et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 (2011).
Stein, E. A. et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N. Engl. J. Med. 366, 1108–1118 (2012).
Mullard, A. Drug makers and NIH team up to find and validate targets. Nat. Rev. Drug Discov. 13, 241–243 (2014).
Lyssenko, V. et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N. Engl. J. Med. 359, 2220–2232 (2008).
Tamaki, M. et al. The diabetes-susceptible gene SLC30A8/ZnT8 regulates hepatic insulin clearance. J. Clin. Invest. 123, 4513–4524 (2013).
Nicolson, T. J. et al. Insulin storage and glucose homeostasis in mice null for the granule zinc transporter ZnT8 and studies of the type 2 diabetes-associated variants. Diabetes 58, 2070–2083 (2009).
Pearson, E. Zinc transport and diabetes risk. Nat. Genet. 46, 323–324 (2014).
Rossetti, L., Smith, D., Shulman, G. I., Papachristou, D. & DeFronzo, R. A. Correction of hyperglycemia with phlorizin normalizes tissue sensitivity to insulin in diabetic rats. J. Clin. Invest. 79, 1510–1515 (1987).
Santer, R. & Calado, J. Familial renal glucosuria and SGLT2: from a mendelian trait to a therapeutic target. Clin. J. Am. Soc. Nephrol. 5, 133–141 (2010).
Sabino-Silva, R. et al. The Na+/glucose cotransporters: from genes to therapy. Braz. J. Med. Biol. Res. 43, 1019–1026 (2010).
Zhang, M. et al. Dapagliflozin treatment for type 2 diabetes: a systematic review and meta-analysis of randomized controlled trials. Diabetes Metab. Res. Rev. 30, 204–221 (2014).
Van Gaal, L. & Scheen, A. Weight management in type 2 diabetes: current and emerging approaches to treatment. Diabetes Care 38, 1161–1172 (2015).
Zhou, K. et al. Clinical and genetic determinants of progression of type 2 diabetes: a DIRECT study. Diabetes Care 37, 718–724 (2014).
Chang, R. L., Xie, L., Xie, L., Bourne, P. E. & Palsson, B. O. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput. Biol. 6, e1000938 (2010).
Mallal, S. et al. HLA-B*5701 screening for hypersensitivity to abacavir. N. Engl. J. Med. 358, 568–579 (2008).
Csermely, P., Korcsmaros, T., Kiss, H. J., London, G. & Nussinov, R. Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review. Pharmacol. Ther. 138, 333–408 (2013).
Taneera, J. et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 16, 122–134 (2012).
Gaulton, K. J. et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 47, 1415–1425 (2015).
Carter, H., Hofree, M. & Ideker, T. Genotype to phenotype via network analysis. Curr. Opin. Genet. Dev. 23, 611–621 (2013).
Cao, C. & Moult, J. GWAS and drug targets. BMC Genomics 15, S5 (2014).
Segre, A. V., Wei, N., Altshuler, D. & Florez, J. C. Pathways targeted by antidiabetes drugs are enriched for multiple genes associated with type 2 diabetes risk. Diabetes 64, 1470–1483 (2015).
Ideker, T. & Krogan, N. J. Differential network biology. Mol. Syst. Biol. 8, 565 (2012).
Greene, C. S. et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015).
Bossi, A. & Lehner, B. Tissue specificity and the human protein interaction network. Mol. Syst. Biol. 5, 260 (2009).
Magger, O., Waldman, Y. Y., Ruppin, E. & Sharan, R. Enhancing the prioritization of disease-causing genes through tissue specific protein interaction networks. PLoS Comput. Biol. 8, e1002690 (2012).
Erler, J. T. & Linding, R. Network medicine strikes a blow against breast cancer. Cell 149, 731–733 (2012).
Collins, F. S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015).
Siva, N. UK gears up to decode 100,000 genomes from NHS patients. Lancet 385, 103–104 (2015).
Koivula, R. W. et al. Discovery of biomarkers for glycaemic deterioration before and after the onset of type 2 diabetes: rationale and design of the epidemiological studies within the IMI DIRECT Consortium. Diabetologia 57, 1132–1142 (2014).
Miller, R. A. et al. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494, 256–260 (2013).
Mulherin, A. J. et al. Mechanisms underlying metformin-induced secretion of glucagon-like peptide-1 from the intestinal L cell. Endocrinology 152, 4610–4619 (2011).
Gribble, F. M. & Reimann, F. Pharmacological modulation of KATP channels. Biochem. Soc. Trans. 30, 333–339 (2002).
Dornhorst, A. Insulinotropic meglitinide analogues. Lancet 358, 1709–1716 (2001).
Yki-Jarvinen, H. Thiazolidinediones. N. Engl. J. Med. 351, 1106–1118 (2004).
Verspohl, E. J. Novel therapeutics for type 2 diabetes: incretin hormone mimetics (glucagon-like peptide-1 receptor agonists) and dipeptidyl peptidase-4 inhibitors. Pharmacol. Ther. 124, 113–138 (2009).
Bailey, C. J. Renal glucose reabsorption inhibitors to treat diabetes. Trends Pharmacol. Sci. 32, 63–71 (2011).
Acknowledgements
E.R.P. holds a Wellcome Trust New Investigator Award 102820/Z/13/Z.
Author information
Authors and Affiliations
Contributions
All authors contributed to all aspects of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Zhou, K., Pedersen, H., Dawed, A. et al. Pharmacogenomics in diabetes mellitus: insights into drug action and drug discovery. Nat Rev Endocrinol 12, 337–346 (2016). https://doi.org/10.1038/nrendo.2016.51
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrendo.2016.51
This article is cited by
-
MIR4532 gene variant rs60432575 influences the expression of KCNJ11 and the sulfonylureas-stimulated insulin secretion
Endocrine (2019)
-
MicroRNA profiling of diabetic atherosclerosis in a rat model
European Journal of Medical Research (2018)
-
Actions of metformin and statins on lipid and glucose metabolism and possible benefit of combination therapy
Cardiovascular Diabetology (2018)
-
Interdisciplinary approach to compensation of hypoglycemia in diabetic patients with chronic heart failure
Heart Failure Reviews (2018)
-
Glucose uptake of the muscle and adipose tissues in diabetes and obesity disease models: evaluation of insulin and β3-adrenergic receptor agonist effects by 18F-FDG
Annals of Nuclear Medicine (2017)