Abstract
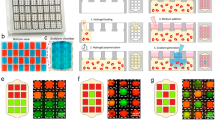
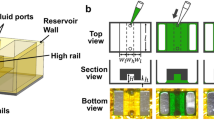
This protocol describes a simple but robust microfluidic assay combining three-dimensional (3D) and two-dimensional (2D) cell culture. The microfluidic platform comprises hydrogel-incorporating chambers between surface-accessible microchannels. By using this platform, well-defined biochemical and biophysical stimuli can be applied to multiple cell types interacting over distances of <1 mm, thereby replicating many aspects of the in vivo microenvironment. Capabilities exist for time-dependent manipulation of flow and concentration gradients as well as high-resolution real-time imaging for observing spatial-temporal single-cell behavior, cell-cell communication, cell-matrix interactions and cell population dynamics. These heterotypic cell type assays can be used to study cell survival, proliferation, migration, morphogenesis and differentiation under controlled conditions. Applications include the study of previously unexplored cellular interactions, and they have already provided new insights into how biochemical and biophysical factors regulate interactions between populations of different cell types. It takes 3 d to fabricate the system and experiments can run for up to several weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Pampaloni, F., Reynaud, E.G. & Stelzer, E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 8, 839–845 (2007).
Even-Ram, S. & Yamada, K.M. Cell migration in 3D matrix. Curr. Opin. Cell Biol. 17, 524–532 (2005).
Feder-Mengus, C., Ghosh, S., Reschner, A., Martin, I. & Spagnoli, G.C. New dimensions in tumor immunology: what does 3D culture reveal? Trends Mol. Med. 14, 333–340 (2008).
Griffith, L.G. & Swartz, M.A. Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 7, 211–224 (2006).
Yamada, K.M. & Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601–610 (2007).
Cukierman, E., Pankov, R. & Yamada, K. Cell interactions with three-dimensional matrices. Curr. Opin. Cell Biol. 14, 633–640 (2002).
Wenger, A. et al. Modulation of in vitro angiogenesis in a three-dimensional spheroidal coculture model for bone tissue engineering. Tissue Eng. 10, 1536–1547 (2004).
Ghajar, C.M., Blevins, K.S., Hughes, C.C.W., George, S.C. & Putnam, A.J. Mesenchymal stem cells enhance angiogenesis in mechanically viable prevascularized tissues via early matrix metalloproteinase upregulation. Tissue Eng. 12, 2875–2888 (2006).
Chung, S., Sudo, R., Vickerman, V., Zervantonakis, I. & Kamm, R. Microfluidic platforms for studies of angiogenesis, cell migration, and cell-cell interactions. Ann. Biomed. Eng. 38, 1164–1177 (2010).
Park, J.W., Vahidi, B., Taylor, A.M., Rhee, S.W. & Jeon, N.L. Microfluidic culture platform for neuroscience research. Nat. Protoc. 1, 2128–2136 (2006).
Paguirigan, A.L. & Beebe, D.J. Protocol for the fabrication of enzymatically crosslinked gelatin microchannels for microfluidic cell culture. Nat. Protoc. 2, 1782–1788 (2007).
Englert, D.L., Manson, M.D. & Jayaraman, A. Investigation of bacterial chemotaxis in flow-based microfluidic devices. Nat. Protoc. 5, 864–872 (2010).
Chung, S., Sudo, R., Zervantonakis, I., Rimchala, T. & Kamm, R. Surface treatment induced three dimensional capillary morphogenesis in a microfluidic platform. Adv. Mater. 21, 4863–4867 (2009).
Chung, S. et al. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip. 9, 269–275 (2009).
Sudo, R. et al. Transport-mediated angiogenesis in 3D epithelial coculture. FASEB J. 23, 2155–2164 (2009).
Vickerman, V., Blundo, J., Chung, S. & Kamm, R. Design, fabrication and implementation of a novel multi parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip. 8, 1468–1477 (2008).
Shin, Y. et al. In vitro 3D collective sprouting angiogenesis under orchestrated ANG-1 and VEGF gradients. Lab Chip. 11, 2175–2181 (2011).
Jeong, G.S. et al. Sprouting angiogenesis under a chemical gradient regulated by interaction with endothelial monolayer in microfluidic platform. Anal Chem. 83, 8454–8459 (2011).
Wan, C., Chung, S. & Kamm, R.D. Differentiation of embryonic stem cells into cardiomyocytes in a compliant microfluidic system. Ann Biomed. Eng. 39, 1840–1847 (2011).
Mack, P.J. et al. Biomechanical regulation of endothelium-dependent events critical for adaptive remodeling. J. Biol. Chem. 284, 8412–8420 (2009).
Kothapalli, C.R. et al. A high-throughput microfluidic assay to study axonal response to growth factor gradients. Lab Chip. 11, 497–507 (2011).
Zervantonakis, I.K. et al. Concentration gradients in microfluidic 3D matrix cell culture systems. Inter. J. Micro-Nano Scale Transport 1, 27–36 (2010).
Amadi, O.C. et al. A low resistance microfluidic system for the creation of stable concentration gradients in a defined 3D microenvironment. Biomed. Microdevices 12, 1027–1041 (2010).
Jeon, J.S., Chung, S., Kamm, R.D. & Charest, J.L. Hot embossing for fabrication of a microfluidic 3D cell culture platform. Biomed.. Microdevices 13, 325–333 (2011).
Zervantonakis, I.K., Kothapalli, C.R., Chung, S., Sudo, R. & Kamm, R.D. Microfluidic devices for studying heterotypic cell-cell interactions and tissue specimen cultures under controlled microenvironments. Biomicrofluidics 5, 13406 (2011).
Qin, D., Xia, Y. & Whitesides, G.M. Soft lithography for micro-and nanoscale patterning. Nat. Protoc. 5, 491–502 (2010).
Jeong, G.S. et al. Microfluidic assay of endothelial cell migration in 3D interpenetrating polymer semi-network HA-Collagen hydrogel. Biomed. Microdevices 13, 717–723 (2011).
Acknowledgements
We acknowledge support to S.C. from the National Research Foundation of Korea (grant no. 2010-0023975), to R.S. from Japan Science and Technology Agency and Japan Society for Promotion of Science (22680037, G2212) and to R.D.K. from the National Science Foundation (CBET-0939511). We thank Y. Kikkawa, Tokyo University of Pharmacy and Life Sciences, for generously providing anti-CD146 antibody.
Author information
Authors and Affiliations
Contributions
Y.S. and S.H. equally contributed to this protocol, designing and carrying out the experiments and writing the paper. J.S.J., K.Y., I.K.Z., R.S. and S.C. designed and carried out the experiments. S.C. is responsible for all the experiments described in this article and preparation of the paper. R.D.K. is responsible for providing guidance for the experiments and for editing the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Shin, Y., Han, S., Jeon, J. et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat Protoc 7, 1247–1259 (2012). https://doi.org/10.1038/nprot.2012.051
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.051
This article is cited by
-
The crosstalk between cholangiocytes and hepatic stellate cells promotes the progression of epithelial-mesenchymal transition and periductal fibrosis during Clonorchis sinensis infection
Parasites & Vectors (2024)
-
Low-cost sheath-less microfluidic impedance cytometry for point-of-care applications
Microfluidics and Nanofluidics (2024)
-
An optofluidic platform for interrogating chemosensory behavior and brainwide neural representation in larval zebrafish
Nature Communications (2023)
-
Evaluating the antioxidant potential of resveratrol-gold nanoparticles in preventing oxidative stress in endothelium on a chip
Scientific Reports (2023)
-
Modeling of three-dimensional innervated epidermal like-layer in a microfluidic chip-based coculture system
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.