Abstract
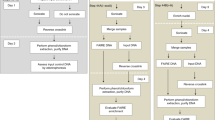
Genomic studies of cell differentiation and function within a whole organism depend on the ability to isolate specific cell types from a tissue, but this is technically difficult. We developed a method called INTACT (isolation of nuclei tagged in specific cell types) that allows affinity-based isolation of nuclei from individual cell types of a tissue, thereby circumventing the problems associated with mechanical purification techniques. In this method nuclei are affinity-labeled through transgenic expression of a biotinylated nuclear envelope protein in the cell type of interest. Total nuclei are isolated from transgenic plants and biotin-labeled nuclei are then purified using streptavidin-coated magnetic beads, without the need for specialized equipment. INTACT gives high yield and purity of nuclei from the desired cell types, which can be used for genome-wide analysis of gene expression and chromatin features. The entire procedure, from nuclei purification through cDNA preparation or chromatin immunoprecipitation (ChIP), can be completed within 2 d. The protocol we present assumes that transgenic lines are already available, and includes procedural details for amplification of cDNA or ChIP DNA prior to microarray or deep sequencing analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Mito, Y., Henikoff, J.G. & Henikoff, S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37, 1090–1097 (2005).
Rao, R.R. & Stice, S.L. Gene expression profiling of embryonic stem cells leads to greater understanding of pluripotency and early developmental events. Biol. Reprod. 71, 1772–1778 (2004).
Irion, S., Nostro, M.C., Kattman, S.J. & Keller, G.M. Directed differentiation of pluripotent stem cells: from developmental biology to therapeutic applications. Cold Spring Harb. Symp. Quant. Biol. 73, 101–110 (2008).
Bhattacharya, B., Puri, S. & Puri, R.K. A review of gene expression profiling of human embryonic stem cell lines and their differentiated progeny. Curr. Stem Cell Res. Ther. 4, 98–106 (2009).
Nakazono, M., Qiu, F., Borsuk, L.A. & Schnable, P.S. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15, 583–596 (2003).
Jiao, Y. et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat. Genet. 41, 258–263 (2009).
Brunskill, E.W. et al. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev. Cell 15, 781–791 (2008).
Birnbaum, K. et al. A gene expression map of the Arabidopsis root. Science 302, 1956–1960 (2003).
Birnbaum, K. et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2, 615–619 (2005).
Gifford, M.L., Dean, A., Gutierrez, R.A., Coruzzi, G.M. & Birnbaum, K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105, 803–808 (2008).
Zhang, Y. et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature 418, 331–335 (2002).
Zhang, C., Barthelson, R.A., Lambert, G.M. & Galbraith, D.W. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 147, 30–40 (2008).
Miller, M.R., Robinson, K.J., Cleary, M.D. & Doe, C.Q. TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat. Methods 6, 439–441 (2009).
Mustroph, A. et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106, 18843–18848 (2009).
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008).
Roy, P.J., Stuart, J.M., Lund, J. & Kim, S.K. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418, 975–979 (2002).
Deal, R.B. & Henikoff, S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040 (2010).
Rose, A. & Meier, I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98, 15377–15382 (2001).
Beckett, D., Kovaleva, E. & Schatz, P.J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 (1999).
Barthelson, R.A., Lambert, G.M., Vanier, C., Lynch, R.M. & Galbraith, D.W. Comparison of the contributions of the nuclear and cytoplasmic compartments to global gene expression in human cells. BMC Genomics 8, 340 (2007).
Jacob, Y., Mongkolsiriwatana, C., Veley, K.M., Kim, S.Y. & Michaels, S.D. The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 144, 1383–1390 (2007).
Meier, I., Xu, X.M., Brkljacic, J., Zhao, Q. & Wang, H.-J. Going green plants' alternative way to position the Ran gradient. J. Microsc. 231, 225–233 (2008).
An, Y.Q. et al. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121 (1996).
Zilberman, D., Coleman-Derr, D., Ballinger, T. & Henikoff, S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129 (2008).
Ruzicka, D.R., Kandasamy, M.K., McKinney, E.C., Burgos-Rivera, B. & Meagher, R.B. The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52, 460–472 (2007).
Masucci, J.D. et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260 (1996).
Gendrel, A.V., Lippman, Z., Martienssen, R. & Colot, V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218 (2005).
Galbraith, D.W. et al. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049–1051 (1983).
Acknowledgements
We thank M. Gehring, F. Steiner and P. Talbert for helpful suggestions on improving the article. This work was supported by funding from the Howard Hughes Medical Institute to S.H. and a Ruth L. Kirschstein Postdoctoral Fellowship from the National Institutes of Health to R.B.D.
Author information
Authors and Affiliations
Contributions
R.B.D. developed the protocol with guidance from S.H. The article was written by R.B.D. and edited by S.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Deal, R., Henikoff, S. The INTACT method for cell type–specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc 6, 56–68 (2011). https://doi.org/10.1038/nprot.2010.175
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.175
This article is cited by
-
Optimization of ATAC-seq in wheat seedling roots using INTACT-isolated nuclei
BMC Plant Biology (2023)
-
Early-life exercise primes the murine neural epigenome to facilitate gene expression and hippocampal memory consolidation
Communications Biology (2023)
-
Lola-I is a promoter pioneer factor that establishes de novo Pol II pausing during development
Nature Communications (2023)
-
The eINTACT method for studying nuclear changes in host plant cells targeted by bacterial effectors in native infection contexts
Nature Protocols (2023)
-
Laser Microdissection of Woody and Suberized Plant Tissues for RNA-Seq Analysis
Molecular Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.