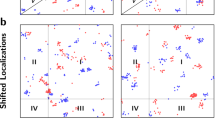
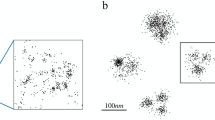
Abstract
Cell function is regulated by the spatiotemporal organization of the signaling machinery, and a key facet of this is molecular clustering. Here, we present a protocol for the analysis of clustering in data generated by 2D single-molecule localization microscopy (SMLM)—for example, photoactivated localization microscopy (PALM) or stochastic optical reconstruction microscopy (STORM). Three features of such data can cause standard cluster analysis approaches to be ineffective: (i) the data take the form of a list of points rather than a pixel array; (ii) there is a non-negligible unclustered background density of points that must be accounted for; and (iii) each localization has an associated uncertainty in regard to its position. These issues are overcome using a Bayesian, model-based approach. Many possible cluster configurations are proposed and scored against a generative model, which assumes Gaussian clusters overlaid on a completely spatially random (CSR) background, before every point is scrambled by its localization precision. We present the process of generating simulated and experimental data that are suitable to our algorithm, the analysis itself, and the extraction and interpretation of key cluster descriptors such as the number of clusters, cluster radii and the number of localizations per cluster. Variations in these descriptors can be interpreted as arising from changes in the organization of the cellular nanoarchitecture. The protocol requires no specific programming ability, and the processing time for one data set, typically containing 30 regions of interest, is ∼18 h; user input takes ∼1 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S.T., Girirajan, T.P.K. & Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. 47, 6172–6176 (2008).
Heilemann, M., van de Linde, S., Mukherjee, A. & Sauer, M. Super-resolution imaging with small organic fluorophores. Angew. Chem. Int. Ed. 48, 6903–6908 (2009).
Kiuchi, T., Higuchi, M., Takamura, A., Maruoka, M. & Watanabe, N. Multitarget super-resolution microscopy with high-density labeling by exchangeable probes. Nat. Methods 12, 743–746 (2015).
Henriques, R. et al. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods 7, 339–340 (2010).
Ovesný, M., Krˇížek, P., Borkovec, J., Švindrych, Z. & Hagen, G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Thompson, R.E., Larson, D.R. & Webb, W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82, 2775–2783 (2002).
Quan, T., Zeng, S. & Huang, Z.-L. Localization capability and limitation of electron-multiplying charge-coupled, scientific complementary metal-oxide semiconductor, and charge-coupled devices for superresolution imaging. J. Biomed. Opt. 15, 066005 (2010).
Rubin-Delanchy, P. et al. Bayesian cluster identification in single-molecule localization microscopy data. Nat. Methods 12, 1072–1076 (2015).
Wolter, S. et al. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods 9, 1040–1041 (2012).
Sage, D. et al. Quantitative evaluation of software packages for single-molecule localization microscopy. Nat. Methods 12, 717–724 (2015).
Williamson, D.J. et al. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat. Immunol. 12, 655–662 (2011).
Sherman, E. et al. Functional nanoscale organization of signaling molecules downstream of the T Cell antigen receptor. Immunity 35, 705–720 (2011).
Garcia-Parajo, M.F., Cambi, A., Torreno-Pina, J.A., Thompson, N. & Jacobson, K. Nanoclustering as a dominant feature of plasma membrane organization. J. Cell Sci. 127, 4995–5005 (2014).
Pageon, S.V. et al. Superresolution microscopy reveals nanometer-scale reorganization of inhibitory natural killer cell receptors upon activation of NKG2D. Sci. Signal. 6, ra62 (2013).
Lin, W.-C. et al. H-Ras forms dimers on membrane surfaces via a protein–protein interface. Proc. Natl. Acad. Sci. USA 111, 2996–3001 (2014).
Lewitzky, M., Simister, P.C. & Feller, S.M. Beyond 'furballs' and 'dumpling soups' – towards a molecular architecture of signaling complexes and networks. FEBS Lett. 586, 2740–2750 (2012).
Kenworthy, A.K. Nanoclusters digitize Ras signalling. Nat. Cell Biol. 9, 875–877 (2007).
Roob, E. III, Trendel, N., Rein ten Wolde, P. & Mugler, A. Cooperative clustering digitizes biochemical signaling and enhances its fidelity. Biophys. J. 110, 1661–1669 (2016).
Stone, M.B. & Veatch, S.L. Steady-state cross-correlations for live two-colour super-resolution localization data sets. Nat. Commun. 6, 7347 (2015).
Simons, K. & Ikonen, E. Functional rafts in cell membranes. Nature 387, 569–572 (1997).
Brown, D.A. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology 21, 430–439 (2006).
Owen, D.M., Williamson, D.J., Magenau, A. & Gaus, K. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat. Commun. 3, 1256 (2012).
Lingwood, D. & Simons, K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010).
Gowrishankar, K. et al. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 149, 1353–1367 (2012).
Goswami, D. et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 135, 1085–1097 (2008).
Köster, D.V. et al. Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc. Natl. Acad. Sci. USA 113, E1645–E1654 (2016).
Ripley, B.D. Modelling spatial patterns. J. R. Stat. Soc. Ser. B Stat. Methodol. 39, 172–192 (1977).
Getis, A. & Franklin, J. Second-order neighborhood analysis of mapped point patterns. Ecology 68, 473–477 (1987).
Owen, D.M. et al. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J. Biophotonics 3, 446–454 (2010).
Lagache, T., Lang, G., Sauvonnet, N. & Olivo-Marin, J.-C. Analysis of the spatial organization of molecules with robust statistics. PLoS One 8, e80914 (2013).
Sengupta, P. et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods 8, 969–975 (2011).
Sengupta, P. & Lippincott-Schwartz, J. Quantitative analysis of photoactivated localization microscopy (PALM) datasets using pair-correlation analysis. BioEssays 34, 396–405 (2012).
Veatch, S.L. et al. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS One 7, e31457 (2012).
Ester, M., Kriegel, H.-P., Sander, J. & Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD-96 Proceedings 226–231 (1996).
Dudok, B. et al. Cell-specific STORM super-resolution imaging reveals nanoscale organization of cannabinoid signaling. Nat. Neurosci. 18, 75–86 (2015).
Levet, F. et al. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods 12, 1065–1071 (2015).
Caetano, F.A. et al. MIiSR: molecular interactions in super-resolution imaging enables the analysis of protein interactions, dynamics and formation of multi-protein structures. PLoS Comput. Biol. 11, e1004634 (2015).
Barna, L. et al. Correlated confocal and super-resolution imaging by VividSTORM. Nat. Protoc. 11, 163–183 (2016).
Malkusch, S. et al. Coordinate-based colocalization analysis of single-molecule localization microscopy data. Histochem. Cell Biol. 137, 1–10 (2012).
Rossy, J., Cohen, E., Gaus, K. & Owen, D.M. Method for co-cluster analysis in multichannel single-molecule localisation data. Histochem. Cell Biol. 141, 605–612 (2014).
Owen, D.M. et al. Quantitative analysis of three-dimensional fluorescence localization microscopy data. Biophys. J. 105, L05–L07 (2013).
Annibale, P., Vanni, S., Scarselli, M., Rothlisberger, U. & Radenovic, A. Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS One 6, e22678 (2011).
Lee, S.-H., Shin, J.Y., Lee, A. & Bustamante, C. Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM). Proc. Natl. Acad. Sci. USA 109, 17436–17441 (2012).
Dempsey, G.T., Vaughan, J.C., Chen, K.H., Bates, M. & Zhuang, X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027–1036 (2011).
Annibale, P., Scarselli, M., Kodiyan, A. & Radenovic, A. Photoactivatable fluorescent protein mEos2 displays repeated photoactivation after a long-lived dark state in the red photoconverted form. J. Phys. Chem. Lett. 1, 1506–1510 (2010).
Gould, T.J., Verkhusha, V.V. & Hess, S.T. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat. Protoc. 4, 291–308 (2009).
van de Linde, S. et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6, 991–1009 (2011).
Sengupta, P., Jovanovic-Talisman, T. & Lippincott-Schwartz, J. Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat. Protoc. 8, 345–354 (2013).
Axelrod, D. Cell surface contacts illuminated by total internal refection fluorescence. J. Cell Biol. 89, 141–145 (1981).
Axelrod, D. Total internal reflection fluorescence microscopy in cell biology. Traffic 2, 764–774 (2001).
Wang, S., Moffitt, J.R., Dempsey, G.T., Xie, X.S. & Zhuang, X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl. Acad. Sci. USA 111, 8452–8457 (2014).
Subach, O.M. et al. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nat. Methods 8, 771–777 (2011).
Brakemann, T. et al. A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching. Nat. Biotechnol. 29, 942–947 (2011).
Shroff, H., Galbraith, C.G., Galbraith, J.A. & Betzig, E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423 (2008).
Ries, J., Kaplan, C., Platonova, E., Eghlidi, H. & Ewers, H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 (2012).
Bromley, S.K. et al. The immunological synapse. Annu. Rev. Immunol. 19, 375–396 (2001).
Rossy, J., Owen, D.M., Williamson, D.J., Yang, Z. & Gaus, K. Conformational states of the kinase Lck regulate clustering in early T cell signaling. Nat. Immunol. 14, 82–89 (2013).
Lillemeier, B.F. et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 (2010).
Acknowledgements
D.M.O. acknowledges funding from the European Research Council (FP7 starter grant 337187) and Marie Curie Career Integration Grant 334303. A.P.C. is funded by Arthritis Research UK grants 19652 and 20525. M.S. was supported by the King's Bioscience Institute and the Guy's and St. Thomas' Charity Prize PhD Programme in Biomedical and Translational Science. We acknowledge the use of the King's College Nikon Imaging Centre (NIC).
Author information
Authors and Affiliations
Contributions
J.G., M.S., C.L.B., L.B., N.A.H., D.M.O. and P.R.-D. developed and tested the protocol. J.G. and P.R.-D. wrote the analysis code. G.L.B., D.J.W. and A.P.C. provided samples. J.G., M.S., G.L.B., D.M.O. and P.R.-D. performed imaging, simulations and analysis. J.G., D.M.O. and P.R.-D. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Performance of the algorithm when applied to simulated data with hard edge clusters
a) Spatial distribution over the 3 x 3 μm ROI. b) Heat map of the scores generated. c) Histogram of the estimated number of clusters for each ROI. d) Histogram of the estimated percentage of localisations found in clusters for each ROI. e) Histogram of the estimated number localisations per cluster. f) Histogram of the standard deviation of the detected clusters. n = 30 simulated ROIs.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 (PDF 256 kb)
Supplementary Software
Implementation of the cluster analysis algorithm, including preprocessing steps, simulation, analysis and postprocessing. (ZIP 10 kb)
Supplementary Data
.csv file containing experimental data from a dSTORM acquisition of ZAP-70 at a T-cell immunological synapse. The file can be opened in ThunderSTORM and used to test the protocol. A 3 x 3 μm region in the center of this cell, containing 4,200 localizations, has a processing time of approximately 2 h and 30 min on a standard desktop computer. (CSV 12482 kb)
Rights and permissions
About this article
Cite this article
Griffié, J., Shannon, M., Bromley, C. et al. A Bayesian cluster analysis method for single-molecule localization microscopy data. Nat Protoc 11, 2499–2514 (2016). https://doi.org/10.1038/nprot.2016.149
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.149
This article is cited by
-
A framework for evaluating the performance of SMLM cluster analysis algorithms
Nature Methods (2023)
-
Unbiased choice of global clustering parameters for single-molecule localization microscopy
Scientific Reports (2022)
-
Correction of multiple-blinking artifacts in photoactivated localization microscopy
Nature Methods (2022)
-
Machine learning for cluster analysis of localization microscopy data
Nature Communications (2020)
-
Super Resolution Network Analysis Defines the Molecular Architecture of Caveolae and Caveolin-1 Scaffolds
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.