Abstract
Conventional methods for preparing small-RNA–seq libraries by adaptor ligation generate a significant amount of adaptor dimer, thereby resulting in wasted sequencing reads. These methods also do not capture small 5′-capped and 5′-triphosphorylated RNAs. The ScriptMiner™ small-RNA–seq library preparation technology overcomes these limitations. ScriptMiner™ small-RNA libraries contain greatly reduced amounts of adaptor-dimer when compared to conventional methods and also generate coverage that is characteristic of the entire small-RNA transcriptome.
Main
The small-RNA transcriptome contains a diverse array of RNAs, such as microRNA (miRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), small interfering RNA (siRNA) and piwi-interacting RNA (piRNA). The biogenesis of these classes of RNAs results in a variety of different modifications at the 5′ end: 5′-monophosphate (5′ pN), 5′-triphosphate (5′pppN) or 5′-cap (GpppN). In addition, the 3′ end of the RNA may be modified to include a 2′-O-methyl, 3′-OH instead of the normal 2′,3′-OH.
Current methods of preparing small-RNA–seq libraries involve ligation-tagging (adaptor-ligation) of the 3′ and 5′ ends of the RNA, reverse transcription of the di-tagged RNA into cDNA, and PCR amplification. However, these methods suffer from two major drawbacks. First, they amplify a significant amount of adaptor-dimer that reduces the efficiency of the 5′ ligation reaction and contaminates the sequencing library, leading to a large number of nonproductive sequencing reads. Second, the conventional methods do not capture small 5′-capped and 5′-triphosphorylated RNAs.
The ScriptMiner™ small-RNA–seq library preparation method significantly reduces the amount of adaptor-dimers in the library and enables the user to capture the entire small-RNA transcriptome, including small 5′-capped and 5′-triphosphorylated RNAs. The result is a more sensitive and comprehensive representation of the small transcriptome in the library.
Overview
Figure 1 presents an overview of the ScriptMiner™ process. Briefly, total RNA or size-selected RNA is tagged at its 3′ end with a preadenylated 3′-adaptor oligonucleotide. A significant proportion of the excess 3′-adaptor oligonucleotide, which can form undesired adaptor-dimers, is then enzymatically removed. The default procedure provides the option to tag only 5′ monophosphorylated RNAs, such as miRNA. An alternative step—treatment with tobacco acid pyrophosphatase (TAP: supplied in the kits)—enables capture of the entire small-RNA transcriptome. The di-tagged RNA is reverse-transcribed into cDNA, and the cDNA amplified by PCR. In addition to amplifying the library, the PCR also incorporates the necessary platform-specific adaptor sequences into the library and adds a barcode (index read) to the library, if desired. The PCR-amplified library is gel-purified, and the extracted small-RNA library is ready for cluster generation before sequencing.
Reduced adaptor-dimer
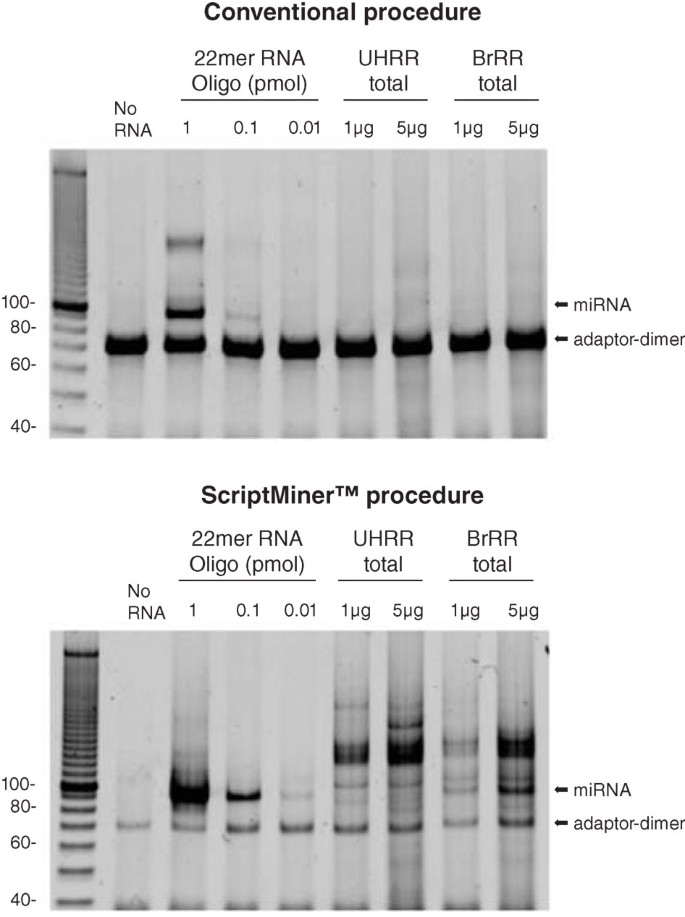
The ScriptMiner™ procedure uses an optimized strategy for degrading excess 3′ adaptor oligonucleotide in order to suppress the formation of undesired adaptor-dimers. Figure 2 shows the greatly reduced level of adaptor-dimer formed by the ScriptMiner™ process compared to a conventional small-RNA–seq library method. By significantly reducing the amount of adaptor-dimer, more of the desired small-RNA transcriptome is amplified by PCR, and less adaptor-dimer contaminates the final sequencing library.
(a) Small-RNA libraries made using a conventional procedure. (b) Small-RNA libraries made using the ScriptMiner™ procedure. Note the significantly reduced level of adaptor oligonucleotide and the enhanced amount of miRNA in the libraries produced by the ScriptMiner™ procedure. The ScriptMiner™ singleplex procedure adds 70 nucleotides to the RNA. 22mer RNA oligo, control RNA oligonucleotide provided in the ScriptMiner™ kits; UHRR, universal human reference RNA; BrRR, human brain reference RNA.
The ScriptMiner ™ method captures the entire small-RNA transcriptome
Following degradation of excess 3′ adaptor oligonucleotide, the user has the option of preparing libraries either from small RNA with a 5′-monophosphate (such as miRNA) or from the entire small-RNA transcriptome, depending on the enzymes chosen in the degradation steps (Fig. 1). By treating the sample with TAP, small RNA with a 5′ monophosphate, a 5′ triphosphate or a 5′ cap will all be included in the library. Table 1 shows libraries prepared from only 5′-monophosphorylated RNA ('No TAP') and from TAP-treated samples. The TAP-treated library produced twice as many aligned reads as the 'No-TAP' library, indicating that the TAP-treated library captured more of the small-RNA transcriptome than conventionally produced ('No-TAP') libraries.
Conclusions
The ScriptMiner™ procedure significantly reduces the amount of adaptor-dimers in small-RNA libraries compared to conventional methods, thereby reducing the number of wasted sequencing reads. ScriptMiner™ libraries can be prepared from the whole small-RNA transcriptome, providing a more detailed picture of the regulatory processes that are mediated by small RNAs within a cell. Current ScriptMiner™ kits permit the preparation of both non-barcoded (singleplex) and barcoded (multiplex) Illumina®-compatible libraries.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pease, J. Small-RNA sequencing libraries with greatly reduced adaptor-dimer background. Nat Methods 8, iii–iv (2011). https://doi.org/10.1038/nmeth.f.336
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.f.336