Abstract
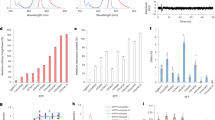
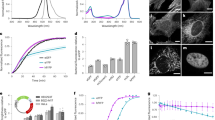
Optimization of autofluorescent proteins by intensity-based screening of bacteria does not necessarily identify the brightest variant for eukaryotes. We report a strategy to screen excited state lifetimes, which identified cyan fluorescent proteins with long fluorescence lifetimes (>3.7 ns) and high quantum yields (>0.8). One variant, mTurquoise, was 1.5-fold brighter than mCerulean in mammalian cells and decayed mono-exponentially, making it an excellent fluorescence resonance energy transfer (FRET) donor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Heim, R. & Tsien, R.Y. Curr. Biol. 6, 178–182 (1996).
Chudakov, D.M., Lukyanov, S. & Lukyanov, K.A. Trends Biotechnol. 23, 605–613 (2005).
Miyawaki, A. Dev. Cell 4, 295–305 (2003).
Kremers, G.J., Goedhart, J., van Munster, E.B. & Gadella, T.W. Jr. Biochemistry 45, 6570–6580 (2006).
Rizzo, M.A., Springer, G.H., Granada, B. & Piston, D.W. Nat. Biotechnol. 22, 445–449 (2004).
van Munster, E.B. & Gadella, T.W. Adv. Biochem. Eng. Biotechnol. 95, 143–175 (2005).
Merzlyak, E.M. et al. Nat. Methods 4, 555–557 (2007).
Mena, M.A., Treynor, T.P., Mayo, S.L. & Daugherty, P.S. Nat. Biotechnol. 24, 1569–1571 (2006).
Nguyen, A.W. & Daugherty, P.S. Nat. Biotechnol. 23, 355–360 (2005).
Chen, Y., Muller, J.D., So, P.T. & Gratton, E. Biophys. J. 77, 553–567 (1999).
Szymczak, A.L. et al. Nat. Biotechnol. 22, 589–594 (2004).
Digman, M.A., Caiolfa, V.R., Zamai, M. & Gratton, E. Biophys. J. 94, L14–L16 (2008).
Pepperkok, R., Squire, A., Geley, S. & Bastiaens, P.I. Curr. Biol. 9, 269–272 (1999).
Verveer, P.J., Wouters, F.S., Reynolds, A.R. & Bastiaens, P.I. Science 290, 1567–1570 (2000).
Yasuda, R. et al. Nat. Neurosci. 9, 283–291 (2006).
Riedl, J. et al. Nat. Methods 5, 605–607 (2008).
Vermeer, J.E., Van Munster, E.B., Vischer, N.O. & Gadella, T.W. Jr. J. Microsc. 214, 190–200 (2004).
Bacia, K. & Schwille, P. Methods 29, 74–85 (2003).
Perroud, T.D., Huang, B. & Zare, R.N. ChemPhysChem 6, 905–912 (2005).
Acknowledgements
We thank I. Elzenaar and M. Adjobo-Hermans for pilot experiments on the 2A co-expression assay and the members of our laboratory for encouraging discussions, J.E.M. Vermeer (University of Amsterdam) for providing plasmid encoding peroxi-YFP with a C-terminal SKL peroxisomal targeting sequence and E.L. Snapp (Albert Einstein College of Medicine) for providing plasmid encoding ER-tdTomato. Part of this work is supported by the EU integrated project on 'Molecular Imaging' (LSHG-CT-2003-503259).
Author information
Authors and Affiliations
Contributions
J.G. and T.W.J.G. designed research; J.G., L.v.W., M.A.H., K.J. and T.W.J.G. conducted experiments; J.G., L.v.W., M.A.H., N.O.E.V. and T.W.J.G. analyzed data; and J.G. and T.W.J.G. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1, Supplementary Note and Supplementary Data (PDF 4861 kb)
Rights and permissions
About this article
Cite this article
Goedhart, J., van Weeren, L., Hink, M. et al. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat Methods 7, 137–139 (2010). https://doi.org/10.1038/nmeth.1415
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1415
This article is cited by
-
A brilliant monomeric red fluorescent protein combining high brightness and fast maturation
Nature Methods (2023)
-
Optimising expression of the large dynamic range FRET pair mNeonGreen and superfolder mTurquoise2ox for use in the Escherichia coli cytoplasm
Scientific Reports (2022)
-
Phasor S-FLIM: a new paradigm for fast and robust spectral fluorescence lifetime imaging
Nature Methods (2021)
-
FUCCI-Red: a single-color cell cycle indicator for fluorescence lifetime imaging
Cellular and Molecular Life Sciences (2021)
-
Multiparameter screening method for developing optimized red-fluorescent proteins
Nature Protocols (2020)