Abstract
DDX41 is a sensor of intracellular double-stranded DNA (dsDNA) in myeloid dendritic cells (mDCs) that triggers a type I interferon response via the signaling adaptor STING. We identified the E3 ligase TRIM21 as a DDX41-interacting protein and found that knockdown of or deficiency in TRIM21 resulted in enhanced type I interferon responses to intracellular dsDNA and DNA viruses. Overexpression of TRIM21 resulted in more degradation of DDX41 and less production of interferon-β (IFN-β) in response to intracellular dsDNA. The SPRY-PRY domain of TRIM21 interacted with the DEADc domain of DDX41. Lys9 and Lys115 of DDX41 were the targets of TRIM21-mediated ubiquitination. TRIM21 is therefore an interferon-inducible E3 ligase that induces the Lys48 (K48)-linked ubiquitination and degradation of DDX41 and negatively regulates the innate immune response to intracellular dsDNA.
Similar content being viewed by others
Main
The innate immune system detects viral infection mainly by sensing viral nucleic acids either in the endosomes or in the cytosol. So far, many sensors of cytosolic double-stranded RNA (dsRNA) have been identified, including RIG-I, Mda5, LGP-2, DDX3, the DDX1-DDX21-DHX36 complex and DHX9, which use the the adaptors MAVS (IPS-1) or TRIF to activate the type I interferon response1,2,3,4,5,6,7. In parallel, many sensors of cytosolic viral DNA have been identified, including AIM2 (refs. 8,9,10,11,12,13,14), DAI (ref. 15), RNA polymerase III–RIG-I (refs. 16,17), IFI16 (ref. 18), the helicase DDX41 (ref. 19) and LRRFIP1 (ref. 20). AIM2 uses the adaptor ASC to activate an inflammasome response; and RNA polymerase III–RIG-I uses MAVS21,22,23, and IFI16 and DDX41 use the signal adaptor STING24,25,26, to activate the type I interferon response.
Uncontrolled sensing of DNA or RNA and excessive production of type I interferon and proinflammatory cytokines have been linked to the development of autoimmune diseases such as systemic lupus erythematosus27. One mechanism developed by the immune system to negatively control the excessive cytokine responses induced by DNA or RNA is direct blockade of the function of the key signaling molecules. For example, NLRX1 (a member of the Nod-like receptor family) inhibits MAVS-mediated interferon-β (IFN-β) responses by disrupting the interaction of RIG-I with MAVS28. Dihydroxyacetone kinase inhibits Mda5-mediated IFN-β responses by binding Mda5 directly29. Another mechanism is enhancement of protein ubiquitination and degradation of key signaling molecules involved in the DNA- or RNA-induced cytokine responses by members of the TRIM family or RING domain–containing family of proteins. Members of the TRIM family (also known as 'RBCC' proteins) contain a RING-finger domain cluster, one or two zinc-binding motifs (called 'B boxes'), a coiled-coil domain and a SPRY domain. The RING domain has E3 ubiquitin ligase activity. TRIM38 (a member of the TRIM family) negatively regulates Toll-like receptor (TLR) signaling by promoting Lys48 (K48)-linked ubiquitination and degradation of TRAF6 (ref. 30). RNF5 (a member of the RING domain–containing family) negatively regulates the production of IFN-β by inducing ubiquitination and degradation of STING, the key adaptor for sensors of cytosolic DNA31. Here we report that the E3 ligase TRIM21 negatively regulated the type I interferon response in myeloid dendritic cells (mDCs) and monocytes that had been induced by cytosolic double-stranded DNA (dsDNA), mainly by promoting the ubiquitination and degradation of DDX41.
Results
Isolation of a DDX41 complex in mDCs
DDX41 is a cytosolic sensor of DNA in mDCs that is critical for triggering STING-dependent type I interferon responses19. To further investigate how DDX41 is regulated in DNA- or DNA virus–induced signaling events, we identified DDX41-interacting proteins by immunoprecipitation with antibody to DDX41 (anti-DDX41) in the mouse mDC line D2SC, followed by protein sequencing by liquid chromatography–mass spectrometry. We obtained approximately 60 unique sequences with four or more 'hits'. DDX41 itself was immunoprecipitated by anti-DDX41, which confirmed the validity of our method for isolating DDX41-binding proteins in mDCs (Supplementary Table 1). We identified the E3 ubiquitin ligase TRIM21, polyubiquitin B, polyubiquitin C and the ubiquitin-like modifier-activating enzyme among the group of DDX41-interacting proteins. Because ubiquitination and members of the TRIM family have been linked to the regulation of innate immune responses, we decided to study the potential function of the E3 ubiquitin ligase TRIM21 in sensing cytosolic DNA.
The role of TRIM21 in sensing cytosolic DNA in mDCs
We first established stable knockdown of TRIM21 and DDX41 mDC cell lines through the use of short hairpin RNA (shRNA), as reported before19. Two distinct TRIM21-targeting shRNAs (Trim21a and Trim21b) produced efficient knockdown of the expression of TRIM21 (Fig. 1a). We stimulated the cells with the synthetic B-form dsDNA poly(dA:dT) and measured the production of IFN-β by the cultured cells by enzyme-linked immunosorbent assay (ELISA). Consistent with published data19, knockdown of DDX41 abrogated the production of IFN-β by mDCs that had been induced by cytosolic DNA. In contrast, knockdown of TRIM21 enhanced the IFN-β production by mDCs up to threefold in response to intracellular DNA (Fig. 1a). Furthermore, overexpression of TRIM21 in mDCs led to lower expression of DDX41 in these mDCs and up to 70% less IFN-β production by mDCs in response to intracellular DNA (Fig. 1b). In contrast, overexpression of TRIM25, the main E3 ligase of the RNA helicase RIG-I, in mDCs did not affect the response to intracellular DNA (Fig. 1b). These data suggested that TRIM21 negatively regulated DDX41 expression and IFN-β production by mDCs in response to intracellular DNA.
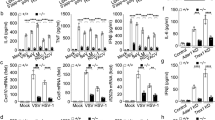
(a) Immunoblot analysis (IB; top) of D2SC cells treated with control shRNA with scrambled sequence (Control) or shRNA targeting TRIM21 (Trim21a and Trim21b) or DDX41 (DDX41) without stimulation; GAPDH serves as a loading control throughout. Below, ELISA of IFN-β in D2SC cells treated with scrambled shRNA and left unstimulated (N-STM) or treated with shRNA as above and then stimulated for 16 h with poly(dA:dT) (0.5 μg/ml). (b) Immunoblot analysis (top) of DDX41, TRIM21 and TRIM25 in D2SC cells (left) or in D2SC cells expressing HA-tagged recombinant full-length TRIM21 (middle) or TRIM25 (right); β-actin serves as a loading control throughout. Below, ELISA of IFN-β in D2SC cells (Control) or in D2SC cells left unstimulated (N-STM) or expressing endogenous or recombinant TRIM21 after 16 h of stimulation with poly(dA:dT) or poly(I:C). (c–e) ELISA of IFN-β in wild-type (WT) or Trim21−/− (KO) BMDCs mock treated (Mock) or stimulated for various times (above graphs) with poly(I:C) (I:C; 5.0 μg/ml) or reovirus (Reo; c); with poly(dA:dT) (AT) or DNA from vaccinia virus (VACA; 0.5 μg/ml each; d); or with adenovirus (Adeno) or HSV-1 (HSV; e). Viruses were used at a multiplicity of infection (MOI) of 10. Each symbol represents an independent experiment; small horizontal lines indicate the average (of triplicates). *P < 0.001 and **P < 0.0002 (Student's t-test). Data are representative of three experiments (error bars a,b), s.d.).
To determine the function of TRIM21 in primary cells, we prepared bone marrow–derived dendritic cells (BMDCs) from wild-type mice and Trim21-deficient mice. We then stimulated those BMDCs for 6, 9 or 20 h with 5′-triphosphate RNA, lipopolysaccharide, the synthetic RNA duplex poly(I:C), poly(dA:dT), DNA from vaccinia virus, the RNA virus reovirus, adenovirus or herpes simplex virus type 1 (HSV-1). Deletion of Trim21 had a small effect on IFN-β production in BMDCs induced by 5′ triphosphate RNA, lipopolysaccharide, poly(I:C) or reovirus (∼20% more production; Fig. 1c and Supplementary Fig. 1), in agreement with a published report noting that TRIM21 negatively regulates the innate immune responses to poly(I:C) and RNA viruses32. In contrast, Trim21-deficient BMDCs produced 1.5- to 4-fold more IFN-β in response to poly(dA:dT), vaccinia-virus DNA, adenovirus or HSV-1 than did wild-type BMDCs (Fig. 1d,e), in agreement with a published report noting that Trim21-deficient cells produce more IFN-β than do wild-type cells in response to HSV-1 (ref. 33). These data indicated a negative role for TRIM21 in the sensing of both intracellular DNA itself and DNA viruses in primary mDCs.
The role of TRIM21 in in vivo host defense
We next evaluated the importance of TRIM21 in vivo in facilitating effective host defense against viral infection. We injected wild-type and Trim21-deficient mice intraperitoneally with HSV-1, then measured viral titers of HSV-1 in peritoneal cells by plaque assay 20 h after infection. We detected significantly less HSV-1 in Trim21-deficient mice than in wild-type mice (titers >90% lower; Fig. 2a). We also infected wild-type mice and Trim21-deficient mice intravenously with HSV-1 or the RNA virus vesicular stomatitis virus and then measured IFN-β, IFN-α and interleukin 6 in serum from infected mice. Trim21-deficient mice produced twofold more type I interferon and interleukin 6 than did wild-type mice, in response to HSV-1 (Fig. 2b,c and Supplementary Fig. 2a). In contrast, deletion of Trim21 had a small effect on the production of type I interferon induced by vesicular stomatitis virus (Fig. 2d and Supplementary Fig. 2b). To further determine the function of TRIM21, we isolated CD11c+ splenocytes (mainly DCs) from wild-type mice and Trim21-deficient mice. Primary CD11c+Trim21-deficient splenocytes produced threefold more IFN-β than did their wild-type counterparts in response to poly(dA:dT) or HSV-1 (Fig. 2e,f). These data indicated an important role for TRIM21 in regulating the innate immune responses to intracellular DNA itself and DNA virus but only a modest (if any) role for TRIM21 in regulating RNA-recognition responses.
(a) Viral titers in wild-type and Trim21−/− mice injected intraperitoneally with HSV-1 (2 × 104 plaque-forming units (PFU)), assessed 20 h later by plaque assay of peritoneal wash fluid. *P < 0.05 (Student's t-test). (b,c) ELISA of IFN-β (b) and interleukin 6 (IL-6; c) in serum from wild-type and Trim21−/− mice 6 h after intravenous infection with HSV-1 (1 × 107 PFU). (d) ELISA of IFN-β in serum from wild-type and Trim21−/− mice 6 h after intravenous infection with vesicular stomatitis virus (VSV; 1 × 107 PFU). (e,f) ELISA of IFN-β in wild-type and Trim21−/− CD11c+ splenocytes after 6 h of stimulation with poly(dA:dT) (0.5 μg/ml; e) or infection with HSV-1 (MOI, 10; f). *P < 0.05 and **P > 0.3 (b–f; Student's t-test). (g) Immunoblot analysis (top) of total DDX41, TRIM21, IRF3, p204 and phosphorylated (p-) IRF3 in lysates of wild-type and Trim21−/− BMDCs stimulated for 1–5 h (above lanes) with poly(dA:dT) (0.5 μg/ml). Below, quantification of bands in the top blot (DDX41); bars correspond to lane markers at top. Data are representative of three experiments (a,g; error bars (a), s.d.) or three independent experiments (b–f; error bars, s.d.).
We next investigated the effect of TRIM21 on the downstream signaling of DDX41 in sensing intracellular DNA in mDCs. We prepared BMDCs from wild-type and Trim21-deficient mice and stimulated the cells with poly(dA:dT). We then prepared total cell extracts and analyzed them by immunoblot. DDX41 expression was higher in Trim21-deficient BMDCs than in wild-type BMDCs (Fig. 2g). TRIM21 expression was upregulated in wild-type BMDCs after stimulation with poly(dA:dT), consistent with the fact that Trim21 is a type I interferon–inducible gene34. IRF3 is a key transcriptional factor downstream of all the sensors of DNA and RNA involved in inducing type I interferon responses. After stimulation with poly(dA:dT), there was more-rapid and robust upregulation of the phosphorylation of IRF3 in Trim21-deficient BMDCs than in wild-type BMDCs (Fig. 2g). In contrast, Trim21 deficiency did not affect the phosphorylation of IRF3 in response to 5′-triphosphate RNA (Supplementary Fig. 2c). In addition, consistent with published data19, the expression of p204 (a type I interferon–inducible sensor of intracellular DNA) was induced by stimulation with dsDNA within 3–5 h in wild-type BMDCs (Fig. 2g). More type I interferon was produced in Trim21-deficient BMDCs than in wild-type BMDCs (data not shown). Consequently, there was more-robust upregulation of p204 in Trim21-deficient BMDCs than in wild-type BMDCs. These data confirmed at the cell-signaling level that TRIM21 negatively controlled the expression of DDX41 protein and its downstream signaling cascade, which was critical for inducing a type I interferon response in mDCs after stimulation with intracellular DNA.
The role of TRIM21 in sensing DNA virus in human cells
Human DDX41 has a critical role in sensing viral DNA in human monocytes19. To determine whether human TRIM21 has a role in sensing DNA itself and DNA viruses, we treated THP-1 human monocyte cells by RNA-mediated interference targeting DDX41 or TRIM21 (or both) and confirmed that expression of the targeted protein was knocked down (Fig. 3a). We then stimulated those THP-1 cells with poly(dA:dT) or HSV-1 and measured IFN-β in the culture supernatants by ELISA. Consistent with published data19, knockdown of DDX41 abrogated the production of IFN-β induced by intracellular DNA or HSV-1. In contrast, knockdown of TRIM21 enhanced the production of IFN-β by THP-1 cells up to threefold in response to intracellular DNA or HSV-1 (Fig. 3b). Further knockdown of TRIM21 in THP-1 cells in which DDX41 had been knocked down had no effect on the production of IFN-β in response to intracellular DNA or HSV-1 (Fig. 3b). We next determined the effect of TRIM21 on downstream signaling by DDX41 in the sensing of intracellular DNA in THP-1 cells. TRIM21 regulated the innate immune response to intracellular DNA in THP-1 cells by a mechanism similar to that used by BMDCs (Supplementary Fig. 3). These data indicated a negative role for TRIM21 in the sensing of both intracellular DNA itself and DNA virus in human monocytes via the targeting of DDX41.
(a) Immunoblot analysis of THP-1 cells treated with scrambled shRNA (sh-control) or with TRIM21-specific siRNA (si-TRIM21) or DDX41-specific shRNA (sh-DDX41) alone (middle) or together (far right). (b) ELISA of IFN-β in THP-1 cells treated with scrambled shRNA and left unstimulated (N-STM) or treated with shRNA or siRNA as in a and stimulated for 16 h with poly(dA:dT) (0.5 μg/ml) or infected for 16 h with HSV-1 (MOI, 10). (c) Immunoblot analysis of wild-type and Trim21−/− BMDCs treated with scrambled or DDX41-specific shRNA (above blot). (d) ELISA of IFN-β in BMDCs treated with scrambled shRNA and left unstimulated (N-STM) or treated with shRNA as in c and stimulated with poly(dA:dT) or HSV-1 as in b. Each symbol represents an independent experiment; small horizontal lines indicate the average (of triplicates). Data are representative of three experiments.
TRIM21 regulates DDX41-mediated innate immune responses
To further determine whether regulation of the innate immune responses to intracellular DNA itself and DNA viruses by TRIM21 was mediated by DDX41, we treated wild-type and Trim21-deficient BMDCs with shRNA targeting DDX41 (Fig. 3c). We stimulated the cells with poly(dA:dT) or HSV-1 and then measured IFN-β in the culture supernatants by ELISA. Knockdown of DDX41 in Trim21-deficient cells abrogated the production of IFN-β induced by intracellular DNA or HSV-1 (Fig. 3d), which confirmed the idea that DDX41 was downstream of TRIM21 in a DNA-sensing pathway and that the regulation of innate immune responses by TRIM21 was mediated by DDX41.
DDX41 binds TRIM21 via the DEADc and SPRY-PRY domains
To characterize the interaction between TRIM21 and DDX41, we assessed resting D2SC mDCs by immunoprecipitation with anti-TRIM21. Anti-TRIM21 did precipitate endogenous DDX41 in resting D2SC mDCs (Fig. 4a). In contrast, anti-TRIM21 did not precipitate DDX21, a member of the helicase family with an important role in sensing intracellular dsRNA6. To determine whether the expression of TRIM21 was regulated by stimulation with DNA, we measured TRIM21 mRNA in D2SC mDCs after stimulation with poly(dA:dT). TRIM21 mRNA was upregulated in D2SC mDCs after stimulation with poly(dA:dT) or poly(I:C), as evaluated by RT-PCR (Fig. 4b), consistent with a published report noting that Trim21 is inducible by type I interferon35.
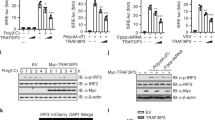
(a) Immunoblot analysis of DDX41 or DDX21 immunoprecipitated (IP) from whole-cell lysates of D2SC cells with anti-DDX21, anti-TRIM21 or anti-DDX41 (above lanes); far left, 10% of input lysate without immunoprecipitation. (b) RT-PCR analysis of TRIM21 and IFN-β in D2SC cells stimulated for 0–8 h (above lanes) with poly(dA:dT) or poly(I:C) (0.5 μg/ml each). (c) Immunoblot analysis (anti-DDX41) of lysates of BMDCs stimulated for 0–4 h (above lanes) with poly(dA:dT) (0.5 μg/ml), before (top) or after (bottom) immunoprecipitation with anti-TRIM21. (d) Full-length TRIM21 and serial truncations of TRIM21 with deletion (Δ) of various domains (left margin); numbers at ends indicate amino acid positions (top). Below, immunoblot analysis of purified Myc-tagged DDX41 with anti-Myc (top blot), and immunoblot analysis (with anti-HA) of purified HA-tagged full-length TRIM21 and TRIM21 truncation mutants alone (middle blot) or after incubation with Myc-tagged DDX41 and immunoprecipitation with anti-Myc (bottom blot). (e) Full-length DDX41 and serial truncations of DDX41 with deletion of various domains (left margin); numbers as in d (top). Below, immunoblot analysis of purified Myc-tagged TRIM21 with anti-Myc (top blot), and immunoblot analysis (with anti-HA) of purified HA-tagged full-length DDX41 and DDX41 truncation mutants alone (middle blot) or after incubation with Myc-tagged TRIM21 and immunoprecipitation with anti-Myc (bottom blot). (f) Confocal microscopy of HEK293T cells cotransfected with expression plasmids for Myc-tagged DDX41 (red) and HA-tagged TRIM21 or TRIM21(ΔPRY) (green), then left unstimulated (N-STM) or stimulated for 4 h with poly(dA:dT) (dA:dT; 0.5 μg/ml); the DNA-intercalating dye DAPI serves as a marker of nuclei. Original magnification, ×100. Below, quantification of the colocalization of TRIM21 or TRIM21(ΔPRY) with DDX41. *P < 0.001 (Student's t-test). Data are representative of three independent experiments (error bars (f), s.d.).
We next investigated the time point at which TRIM21 operated during assembly of the DDX41-DNA signaling complex. We stimulated BMDCs for 0, 1, 2 or 4 h with poly(dA:dT), followed by immunoprecipitation with anti-TRIM21 and immunoblot analysis of DDX41. TRIM21 formed a complex with DDX41 in resting BMDCs, and TRIM21 dissociated from DDX41 after 4 h of stimulation with poly(dA:dT) (Fig. 4c). These data indicated that TRIM21 regulated DDX41 at both the steady state and the early stage of stimulation with dsDNA.
To map the binding sites between DDX41 and TRIM21, we analyzed the interactions between Myc-tagged recombinant DDX41 and hemagglutinin (HA)-tagged recombinant full-length TRIM21 and truncation mutants of TRIM21. Both recombinant full-length TRIM21 and the carboxy-terminal SPRY-PRY domains of TRIM21 bound DDX41 (Fig. 4d). The DEADc domain of DDX41 bound TRIM21 (Fig. 4e).
Because an antibody to DDX41 for cell staining is not available, we expressed Myc-tagged DDX41 together with HA-tagged full-length TRIM21 or TRIM21 with truncation of the carboxyl terminus (TRIM21(ΔPRY), which lacks the DDX41-binding site) in HEK293T human embryonic kidney cells to determine their subcellular localization. Immunofluorescence imaging showed more colocalization of full-length TRIM21 and DDX41 without DNA stimulation (64.15%) than after 4 h of stimulation with DNA (4.37%; Fig. 4f). In contrast, TRIM21(ΔPRY) did not localize together with DDX41 (<1.5%). These data suggested that TRIM21 and DDX41 localized together in the cytosol under resting conditions and that TRIM21 dissociated from DDX41 after stimulation with dsDNA.
TRIM21 induces DDX41 ubiquitination by K48 linkage
To investigate whether DDX41 undergoes protein ubiquitination and degradation, we transfected HEK293T cells with an expression vector for HA-tagged DDX41 and detected HA-tagged DDX41 by immunoblot analysis. DDX41 underwent robust ubiquitination and degradation (Fig. 5a). Overexpression of Myc-tagged TRIM21 together with HA-tagged DDX41 in HEK293T cells enhanced the ubiquitination of DDX41 (Fig. 5b).
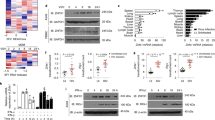
(a) Immunoblot analysis of the ubiquitination (Ub) and abundance of HA-tagged DDX41 in HEK293T cells transfected with expression vector for HA-tagged DDX41 (HA-DDX41) or empty vector (Control). (b) Immunoblot analysis of HA-tagged DDX41 (top blot), the ubiquitination of DDX41 (second blot) and Myc-tagged TRIM21 (third blot) in HEK293T cells cotransfected with expression vector for HA-tagged DDX41 and with empty vector or expression vector for Myc-tagged TRIM21 (above lanes). (c) Immunoblot analysis (with anti-HA) of the abundance (top), total ubiquitination (second blot), K48-linked ubiquitination (third blot) and K63-linked ubiquitination (fourth blot) of HA-tagged DDX41 in L929 cells transfected with empty vector or expression vector for Myc-tagged TRIM21 or TRIM21(ΔRING) and stimulated for 3 h with poly(dA:dT) (0.5 μg/ml), assessed after immunoprecipitation with anti-HA; and immunoblot analysis whole-cell lysates with anti-Myc (bottom). (d) Immunoblot analysis of TRIM21 in wild-type and Trim21−/− BMDCs (top), and of the abundance (second blot), total ubiquitination (third blot) and K48-mediated ubiquitination (bottom panel) of DDX41 in those cells, stimulated as in c, assessed after immunoprecipitation with anti-DDX41. Data are representative of three experiments.
To determine whether the ubiquitination of DDX41 was dependent on the RING domain of TRIM21, we transfected the mouse fibroblast cell line L929 to express HA-tagged DDX41 and either Myc-tagged full-length TRIM21 or truncated TRIM21 that lacked the RING domain (TRIM21(ΔRING)). We stimulated the cells for 3 h with poly(dA:dT), then prepared cell lysates and incubated them for 5 min at 90 °C with 1% SDS to disrupt protein-protein interactions, followed by immunoprecipitation of HA-tagged DDX41. Immunoblot analysis of hemagglutinin or ubiquitin demonstrated that the ubiquitination of DDX41 was strongly enhanced by overexpression of TRIM21 but not by overexpression of TRIM21(ΔRING) (Fig. 5c). Immunoblot analysis of K48-linked or K63-linked ubiquitin further demonstrated that TRIM21 induced ubiquitination of DDX41 by K48-mediated linkage (Fig. 5c).
To determine whether TRIM21 was the main E3 ubiquitin ligase for the ubiquitination of DDX41, we stimulated wild-type and Trim21-deficient BMDCs for 3 h with poly(dA:dT), then prepared cell lysates and analyzed the ubiquitination of DDX41. We detected DDX41 ubiquitination via K48-mediated linkage in wild-type BMDCs but not in Trim21-deficient BMDCs (Fig. 5d). This indicated an exclusive role for TRIM21 in the ubiquitination of DDX41, at least after stimulation with poly(dA:dT).
To determine whether TRIM21 directly modified DDX41, we reconstituted the DDX41-ubiquitination reaction in vitro. We prepared various combinations of recombinant glutathione S-transferase (GST)-tagged DDX41, recombinant TRIM21, purified E2 enzymes (HbcH2, HbcH3, HbcH5a, HbcH5b, HbcH5c, HbcH6, HbcH7, HbcH8, HbcH10 and HbcH13) and a mixture of E1 plus ubiquitin plus ATP, then incubated those preparations for 2 h at 30 °C, followed by treatment for 5 min at 90 °C with 1% SDS. We immunoprecipitated GST-tagged DDX41 with glutathione-Sepharose beads, followed by immunoblot analysis. We observed degradation of DDX41 (Fig. 6a) and ubiquitination of DDX41 (Fig. 6b) only when TRIM21, the mixture of E1 plus ubiquitin plus ATP, and one of the E2 enzymes were present. We found that the ubiquitination of DDX41 was mediated by K48 linkage (Fig. 6c) but not by K63 linkage (data not shown). Together these data indicated that TRIM21 induced the ubiquitination of DDX41 by K48-mediated linkage.
Immunoblot analysis of the degradation (a), total ubiquitination (b) and K48-mediated ubiquitination (c) of GST-tagged DDX41, assessed by in vitro ubiquitination assay with various combinations (above lanes) of a mixture of E1 plus ubiquitin plus ATP (E1-Ub-ATP), TRIM21 (E3 TRIM21), GST-tagged DDX41 (GST-DDX41), GST and purified E2 enzymes (HbcH; numbers above lanes indicate which enzyme), after immunoprecipitation with anti-GST. Data are representative of three experiments.
TRIM21 does not ubiquitinate other sensors of viral DNA
We investigated whether other sensors of viral nucleic acids were regulated by TRIM21. For this, we overexpressed HA-tagged TRIM21 in THP-1 cells and found that this led to lower expression of DDX41 but had no effect on expression of the sensors IFI16, RIG-I, Mda5 or IRF3 (Supplementary Fig. 4). We next determined whether other reported sensors of viral dsDNA were ubiquitinated by TRIM21. We stimulated wild-type or Trim21-deficient BMDCs for 4 h with poly(dA:dT), then analyzed total cell extracts by immunoblot. There was little difference between wild-type and Trim21-deficient BMDCs in the expression of LRRFIP1, p204 or DAI (Supplementary Fig. 5). Next we stimulated those cells for 3 h with poly(dA:dT), followed by immunoprecipitation with anti-LRRFIP1, anti-p204 or anti-DAI. Anti-LRRFIP1, anti-p204 or anti-DAI did precipitate these sensors of viral DNA, which confirmed the validity of our method for the immunoprecipitation of endogenous protein; however, we detected no ubiquitination of these proteins (Supplementary Fig. 6). Furthermore, we detected no difference between wild-type and Trim21-deficient BMDCs in the ubiquitination of LRRFIP1, p204 or DAI. These data indicated that TRIM21 had no role in regulating the expression of LRRFIP1, p204 and DAI by ubiquitination or degradation.
Lys9 and Lys115 ubiquitination sites of DDX41
DDX41 contains 50 lysine residues. Eight of these (Lys9, Lys56, Lys95, Lys97, Lys111, Lys115, Lys597 and Lys604) were predicted to be possible ubiquitination sites by the UbPred program that predicts such sites (Supplementary Table 2). To determine the DDX41-ubiquitination sites, we replaced each of the eight DDX41 lysine residues noted above individually with arginine. We expressed HA-tagged wild-type and mutant DDX41 in L929 cells (Fig. 7a). We stimulated those cells for 3 h with poly(dA:dT), then treated cell lysates for 5 min at 90 °C with 1% SDS. Immunoprecipitation with anti-hemagglutinin and immunoblot analysis of ubiquitin demonstrated that the K9R or K115R substitution partially blocked ubiquitination of DDX41 via K48-mediated linkage (Fig. 7a). We next generated a mutant of DDX41 with substitution of both K9R and K115R and found that the double substitution completely blocked the ubiquitination of DDX41 (Fig. 7a). These data indicated that Lys9 and Lys115 were the ubiquitination sites of DDX41.
(a) Immunoblot analysis of the expression (top), total ubiquitination (third blot) and K48-linked ubiquitination (bottom) of wild-type DDX41 or DDX41 mutants in HEK293T cells transfected with empty vector (Control) or expression vector for HA-tagged wild-type DDX41 (WT) or DDX41 mutants (above lanes), followed by immunoprecipitation anti-HA. (b) Activation of the Ifnb promoter in L929 cells transfected with an Ifnb luciferase reporter, plus expression vector (each 10 ng) for wild-type DDX41 or various DDX41 mutants alone (above) or expression vector for TRIM21 plus wild-type DDX41 or DDX41 mutants (below); results are presented relative to those of renilla luciferase (cotransfected as an internal control), set as 1. *P < 0.001 (Student's t-test). Data are representative of three independent experiments (error bars (b), s.d.).
Through the use of a luciferase reporter assay of the Ifnb promoter, established in the L929 cell line, we found that overexpression of DDX41 enhanced the activity of the Ifnb promoter in response to stimulation with poly(dA:dT). Overexpression of mutant DDX41 with the K9R or K115R substitution or both the K9R and K115R substitutions further enhanced the activity of the Ifnb promoter (Fig. 7b), which confirmed our proposal that Lys9 and Lys115 were important sites for the ubiquitination and degradation of DDX41 at the functional level. In addition, overexpression of TRIM21 inhibited the activity of the Ifnb promoter in L929 cells expressing wild-type DDX41 or mutant DDX41 with the K56R, K95R, K97R, K111R, K597R or K604R substitution. However, overexpression of TRIM21 did not inhibit the activity of the Ifnb promoter in L929 cells expressing mutant DDX41 with the K9R or K115R substitution or both the K9R and K115R substitutions (Fig. 7b). These data indicated that Lys9 and Lys115 were important sites for TRIM21-mediated ubiquitination and regulation of DDX41.
Discussion
Protein ubiquitination is a general mechanism that controls a large number of cellular processes, including protein degradation, DNA repair, chromatin remodeling, cell-cycle regulation, endocytosis and kinase signaling pathways35. Protein ubiquitination is catalyzed by the sequential action of three enzymes: the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2) and the ubiquitin ligase enzyme (E3). Members of the TRIM family have E3 ubiquitin ligase activity. In this study, we discovered that TRIM21 induced ubiquitination and subsequent degradation of DDX41, a sensor of intracellular dsDNA, in mDCs and monocytes. We noted the following structural and functional features of TRIM21: it bound the DDX41 DEADc domain via the carboxy-terminal SPRY-PRY domain; it ubiquitinated DDX41 by K48-mediated linkage for protein degradation but not by K63-mediated linkage for facilitating cell signaling; it ubiquitinated DDX41 at both Lys9 and Lys115; and it was upregulated in mDCs after stimulation with intracellular DNA, consistent with the fact that TRIM21 is encoded by a type I interferon–inducible gene, which links it to the negative feedback regulation of type I interferon responses induced by cytosolic DNA.
Studies have demonstrated that members of the TRIM family can induce ubiquitination by K63-mediated linkage or K48-mediated linkage. Although the former facilitates a protein-protein interaction that is critical for the regulation of cell signaling, the latter permits protein degradation via proteasome-mediated proteolysis. It has been shown that TRIM25 induces ubiquitination of the amino-terminal caspase-recruitment domains of RIG-I by K63-mediated linkage, which is essential for the triggering of type I interferon responses by RIG-I to dsRNA36,37. TRIM56 induces the ubiquitination of STING by K63-mediated linkage, which permits STING, the key adaptor for sensors of DNA, to undergo dimerization and to recruit TBK1 for the initiation of an IFN-β response to intracellular DNA38. In contrast, TRIM38 induces the ubiquitination of TRAF6 by K48-mediated linkage and degradation of TRAF6 in response to the stimulation of macrophages with poly(I:C) and blocks activation of the transcription factor NF-κB and mitogen-activated protein kinases, which subsequently negatively regulates the production of proinflammatory cytokines30.
Activation of signaling by sensors of viral nucleic acids in cells of the innate immune system is important for the elimination of invading microorganisms39,40. However, uncontrolled production of type I interferon induced by DNA and RNA may lead to autoimmune and inflammatory diseases, such as systemic lupus erythematosus. In this context, TRIM21 has been called an 'autoantigen' (RO52 or SS-A) in patients with Sjögren's syndrome and in patients with systemic lupus erythematosus25. The identification of TRIM21 as a negative regulator of sensors of intracellular dsDNA will have important implications for the understanding of not only antiviral innate immunity but also the pathogenesis of human autoimmune diseases such as systemic lupus erythematosus.
Methods
Mice.
Trim21-deficient mice were from Jackson Laboratory. Primary bone marrow was collected from wild-type and Trim21-deficient mice for BMDCs. Spleens were collected from wild-type and Trim21-deficient mice, and single-cell suspensions of splenocytes were sorted for the purification of CD11+ splenocytes. Animals were housed in specific pathogen–free barrier facilities. All experiments were done according to institutional guidelines at the UT MD Anderson Cancer Center.
Reagents.
The 5′ triphosphate RNA, poly(I:C) and lipopolysaccharide were from Invivogen. Lipofectamine 2000 was from Invitrogen. Poly(dA:dT) and the proteasome inhibitor MG132 were from Sigma. DNA from vaccinia virus (sequence, ref. 18) was prepared by Sigma. The following antibodies were used for immunoprecipitation: anti-DDX41 (sc-166225; C3; Santa Cruz), anti-TRIM21 (sc-21367; M-20; Santa Cruz), anti-DDX21 (NB100-41437; Novus Biologicals), anti-DAI (PRS4401; Sigma), anti-IFI16-p204 (I1659; Sigma) and anti-LRRFIP1 (A303-078A; Bethyl). The following antibodies were used for immunoblot analysis: anti-DDX41 (SAB2100554; Sigma), anti-TRIM21 (sc-21367; M-20; Santa Cruz), anti-hTRIM21 (AB4146, Millipore), anti-IFI-204 (sc-13367; M-15; Santa Cruz), anti-DAI (PRS4401; Sigma), anti-IFI16-p204 (I1659; Sigma), anti-RIG-I (3743S; Cell Signaling), anti-Mda5 (5321S; Cell Signaling); anti-TRIM25 (A301-858A; Bethyl), anti-LRRFIP1 (A303-078A; Bethyl), anti-IRF3 (sc-9082; FL-425; Santa Cruz), antibody to phosphorylated IRF3 (4947; Cell Signaling), anti-ubiquitin (sc-8017; Santa Cruz), K63-specific anti-ubiquitin (05-1313; Millipore), K48-specific anti-ubiquitin (05-1307; Millipore), anti-GAPDH (G9295; Sigma), anti-HA (H6533; Sigma), anti-Myc (ab1326; Abcam) and anti-GST (ab58626; Abcam). The following antibodies were used for confocal microscopy: Alexa Fluor 555–anti-Myc (3756; Cell Signaling) and Alexa Fluor 488–anti-HA (2350; Cell Signaling). Anti-HA and anti-Myc beads were from Sigma. NeutAvidin-beads were from Pierce. Glutathione-Sepharose was from GE Healthcare. Lentiviral vectors for shRNA were as follows (all from Open Biosystems): TRIM21 (RMM4534-NM_001082552; clone TRCN 0000040689 (Trim21a) and clone TRCN 0000040688 (Trim21b); DDX41 (RMM4534-NM_134059; clone TRCN 0000104013). GST (NBP1-30259) and GST-tagged DDX41 (H00051428-p01) were from Novus Biologicals. TRIM21 (PRO-328) was from ProSpec. The Ubiquitin Thioester/Conjugation Initiation kit (K-995) and UbcH Enzyme Set (K-980) were from BostonBiochem. The IFN-β ELISA kit was from PBL InterferonSource. The Dual-Luciferase Reporter Assay System (E1910) was from Promega.
Luciferase reporter gene assay.
L929 cells were seeded on 48-well plates (1 × 105 cells per well), then transfected with reporter vectors for Ifnb–firefly luciferase (200 ng) and renilla luciferase (1 ng) plus expression vector for wild-type DDX41 or DDX41 mutants (100 ng) with or without expression vector for TRIM21 (100 ng). Empty control vector was added so that a total of 450 ng of vector DNA was transfected into each well of cells. At 24 h after transfection, cells were stimulated with 0.5 μg/ml poly(dA:dT) delivered by Lipofectamine 2000. Cells were collected after 6 h of stimulation. Luciferase activity in total cell lysates was detected by Dual-Luciferase Reporter Assay.
RNA-mediated interference.
Culture of D2SC mDCs and RNA-mediated interference were done as described6.
Viral titer assay.
HSV-1 infection and viral titer assay were done as described41.
THP-1 cell culture and lentiviral infection.
THP-1 cells were maintained in RPMI-1640 medium containing 10% heat-inactivated FCS and 1% penicillin-streptomycin (Invitrogen-Gibco). THP-1 cells were infected with a pLKO.1 lentiviral vector carrying a target gene sequence or a scrambled shRNA (Open Biosystems). After 24 h of culture, cells were selected by the addition of puromycin (2 ng/ml) to the medium. Cells were stimulated with DNA (0.5 μg/ml) delivered by Lipofectamine 2000 or HSV-1 at an MOI of 10. The knockdown efficiency was detected with immunoblot analysis.
In vitro pull-down and immunoblot analysis.
For the preparation of purified DDX41 and TRIM21, HEK293T cells were transfected with an expression plasmid encoding full-length or truncated versions of HA- or Myc-tagged DDX41 or TRIM21. Lysates were prepared from the transfected cells, followed by incubation with anti-HA or anti-Myc beads. Proteins were eluted from the beads after beads were washed six times with PBS. For precipitation with anti-HA or anti-Myc beads, purified HA-tagged wild-type DDX41 or truncations of DDX41 were incubated for 1 h with purified Myc-tagged TRIM21 or purified HA-tagged TRIM21 or truncations of TRIM21 were incubated for 1 h with purified Myc-tagged DDX41. Beads were added; after 1 h of incubation, the bound complexes were pelleted by centrifugation. Proteins and beads were analyzed by immunoblot analysis with anti-HA or anti-Myc.
RT-PCR.
Total RNA was isolated from cells with an RNeasy kit (Qiagen). Trace amounts of DNA in the RNA extract were digested with DNase (Qiagen). Then, cDNA was prepared with SuperScript II (Invitrogen) and random hexamer primers. The following primers were used for PCR: TRIM21, 5′-CCATGGTGGAGCCTATGAGT-3′ (forward) and 5′-GGTGAAGCTTCTCTCCATGC-3′ (reverse); IFN-β, 5′-CCCTATGGAGATGACGGAGA-3′ (forward) and 5′-TCCCACGTCAATCTTTCCTC-3′ (reverse); and GAPDH, 5′-ACCCAGAAGACTGTGGATGG-3′ (forward) and 5′-CAGTGAGCTTCCCGTTCAG-3′ (reverse).
Ubiquitination.
HEK293T cells were transfected with expression plasmid encoding HA-tagged full-length DDX41 and with or without coexpression of Myc-tagged TRIM21. At 24 h after transfection, cells were stimulated for 3 h with 0.5 μg/ml poly(dA:dT) delivered by Lipofectamine 2000 and 25 μM MG132 and then were collected for analysis. In other experiments, DCs derived with the cytokine GM-CSF were stimulated for 3 h with 0.5 μg/ml of poly(dA:dT) delivered by Lipofectamine 2000 and 25 μM MG132 and then were collected for analysis. Extracts were analyzed by immunoblot or were immunoprecipitated with anti-HA beads or anti-DDX41 and then analyzed by immunoblot.
In vitro ubiquitination assay.
GST-tagged DDX41 (100 ng) and TRIM21 derived from Escherichia coli (100 ng) were incubated with E1 (100 ng), E2 (500 ng) and ubiquitin (2.5 μg) in 50 μl ubiquitination assay buffer (50 mm Tris-HCl, 2.5 mm MgCl2, 0.5 mm DTT and 2 mm ATP). Reactions were incubated for 2 h at 30 °C. Proteins were immunoprecipitated with anti-GST beads and then analyzed by immunoblot.
Confocal microscopy.
HEK293T cells were cotransfected with expression plasmids for HA-tagged TRIM21 or TRIM21(ΔPRY) and Myc-tagged DDX41. After 24 h, cells were stimulated for 4 h with 0.5 μg/ml poly(dA:dT) or left unstimulated. Cells were then fixed in 4% paraformaldehyde and permeabilized with 0.1% saponin, then blocked for 30 min with 10% goat serum, incubated overnight at 4 °C with Alexa Fluor 555–anti-Myc and Alexa Fluor 488–anti-HA and then examined with confocal microscopy. Images of 'zoomed' single cells were quantified with Leica Confocal Software.
References
Yoneyama, M. et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 (2004).
Yoneyama, M. et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175, 2851–2858 (2005).
Kato, H. et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006).
Myong, S. et al. Cytosolic viral sensor RIG-I is a 5′ triphosphate-dependent translocase on double-stranded RNA. Science 323, 1070–1074 (2009).
Oshiumi, H. & Sakai, K. Matsumoto, M. & Seya, T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur. J. Immunol. 40, 940–948 (2010).
Zhang, Z. et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 34, 866–878 (2011).
Zhang, Z., Yuan, B., Lu, N., Facchinetti, V. & Liu, Y.-J. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J. Immunol. 187, 4501–4508 (2011).
Roberts, T.L. et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323, 1057–1060 (2009).
Hornung, V. et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 (2009).
Fernandes-Alnemri, T.Yu., Datta, J.W., Wu, P.J. & Alnemri, E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 (2009).
Bürckstümmer, T. et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10, 266–272 (2009).
Rathinam, V.A. et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402 (2010).
Fernandes-Alnemri, T. et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11, 385–393 (2010).
Jones, J.W. et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. USA 107, 9771–9776 (2010).
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007).
Chiu, Y.H., Macmillan, J.B. & Chen, Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 (2009).
Ablasser, A. et al. RIG-I-dependent sensing of polydA:dT through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10, 1065–1072 (2009).
Unterholzner, L. et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 (2010).
Zhang, Z. et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 (2011).
Yang, P. et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 11, 487–494 (2010).
Kawai, T. et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005).
Seth, R.B., Sun, L., Ea, C.K. & Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 (2005).
Xu, L.G. et al. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 (2005).
Ishikawa, H., Ma, Z. & Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009).
Ishikawa, H. & Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signaling. Nature 455, 674–678 (2008).
Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008).
Münz, C., Lünemann, J.D., Getts, M.T. & Miller, S.D. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat. Rev. Immunol. 9, 246–258 (2009).
Moore, C.B. et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451, 573–577 (2008).
Diao, F. et al. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc. Natl. Acad. Sci. USA 104, 11706–11711 (2007).
Zhao, W., Wang, L., Zhang, M., Yuan, C. & Gao, C. E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated Factor 6 in macrophages. J. Immunol. 188, 2567–2574 (2012).
Zhong, B. et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30, 397–407 (2009).
Higgs, R. et al. The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181, 1780–1786 (2008).
Espinosa, A. et al. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J. Exp. Med. 206, 1661–1671 (2009).
Yoshimi, R. et al. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-κB-dependent cytokine expression in fibroblasts. J. Immunol. 182, 7527–7538 (2009).
Reyes-Turcu, F.E., Ventii, K.H. & Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 (2009).
Gack, M.U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007).
Zeng, W. et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141, 315–330 (2010).
Tsuchida, T. et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33, 765–776 (2010).
Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 449, 819–826 (2007).
Iwasaki, A. & Medzhitov, R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5, 987–995 (2004).
Fujioka, N. et al. Interleukin-18 protects mice against acute herpes simplex virus type 1 infection. J. Virol. 73, 2401–2409 (1999).
Acknowledgements
We thank C. Harrod and L. Snipes for critical reading, and all colleagues in our laboratory at Baylor Institute of Immunology.
Author information
Authors and Affiliations
Contributions
Z.Z. designed and did most of the experiments; M.B., N.L., L.W. and B.Y. helped with the experiments; Z.Z. and Y.-J.L. wrote the manuscript; and Y.-J.L. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Tables 1 and 2 (PDF 239 kb)
Rights and permissions
About this article
Cite this article
Zhang, Z., Bao, M., Lu, N. et al. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol 14, 172–178 (2013). https://doi.org/10.1038/ni.2492
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2492
This article is cited by
-
Cytosolic DNA sensors in neurodegenerative diseases: from physiological defenders to pathological culprits
EMBO Molecular Medicine (2024)
-
TRIM21 inhibits irradiation-induced mitochondrial DNA release and impairs antitumour immunity in nasopharyngeal carcinoma tumour models
Nature Communications (2023)
-
The TRIM21-FOXD1-BCL-2 axis underlies hyperglycaemic cell death and diabetic tissue damage
Cell Death & Disease (2023)
-
AGO4 suppresses tumor growth by modulating autophagy and apoptosis via enhancing TRIM21-mediated ubiquitination of GRP78 in a p53-independent manner
Oncogene (2023)
-
Mutual regulation between TRIM21 and TRIM8 via K48-linked ubiquitination
Oncogene (2023)