Abstract
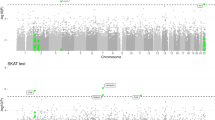
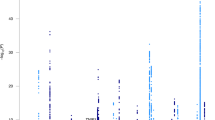
We sought to identify new susceptibility loci for Alzheimer's disease through a staged association study (GERAD+) and by testing suggestive loci reported by the Alzheimer's Disease Genetic Consortium (ADGC) in a companion paper. We undertook a combined analysis of four genome-wide association datasets (stage 1) and identified ten newly associated variants with P ≤ 1 × 10−5. We tested these variants for association in an independent sample (stage 2). Three SNPs at two loci replicated and showed evidence for association in a further sample (stage 3). Meta-analyses of all data provided compelling evidence that ABCA7 (rs3764650, meta P = 4.5 × 10−17; including ADGC data, meta P = 5.0 × 10−21) and the MS4A gene cluster (rs610932, meta P = 1.8 × 10−14; including ADGC data, meta P = 1.2 × 10−16) are new Alzheimer's disease susceptibility loci. We also found independent evidence for association for three loci reported by the ADGC, which, when combined, showed genome-wide significance: CD2AP (GERAD+, P = 8.0 × 10−4; including ADGC data, meta P = 8.6 × 10−9), CD33 (GERAD+, P = 2.2 × 10−4; including ADGC data, meta P = 1.6 × 10−9) and EPHA1 (GERAD+, P = 3.4 × 10−4; including ADGC data, meta P = 6.0 × 10−10).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Gatz, M. et al. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 63, 168–174 (2006).
Saunders, A.M. et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology 43, 1467–1472 (1993).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 41, 1088–1093 (2009).
Lambert, J.C. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 41, 1094–1099 (2009).
Corneveaux, J.J. et al. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum. Mol. Genet. 19, 3295–3301 (2010).
Zhang, Q. et al. Complement receptor 1 polymorphisms and risk of late onset Alzheimer's disease. Brain Res. 1348, 216–221 (2010).
Carrasquillo, M.M. et al. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch. Neurol. 67, 961–964 (2010).
Jun, G. et al. Meta-analysis confirms CR1, CLU, and PICALM as Alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch. Neurol. 67, 1473–1484 (2010).
Kamboh, M.I. et al. Association of CLU and PICALM variants with Alzheimer's disease. Neurobiol. Aging published online, doi:10.1016/j.neurobiolaging.2010.04.015 (4 June 2010).
Biffi, A. et al. Genetic variation and neuroimaging measures in Alzheimer disease. Arch. Neurol. 67, 677–685 (2010).
Seshadri, S. et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. J. Am. Med. Assoc. 303, 1832–1840 (2010).
Naj, A.C. et al. Common variants in MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. advance online publication, doi:10.1038/ng.801 (3 April 2011).
Reiman, E.M. et al. GAB2 alleles modify Alzheimer's risk in APOE ɛ4 carriers. Neuron 54, 713–720 (2007).
Petersen, R.C. et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 74, 201–209 (2010).
Pe'er, I., Yelensky, R., Altshuler, D. & Daly, M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32, 381–385 (2008).
Adzhubei, I.A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Stranger, B.E. et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 1, e78 (2005).
Gibbs, J.R. et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 6, e1000952 (2010).
Bertram, L. et al. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am. J. Hum. Genet. 83, 623–632 (2008).
Kim, W.S., Weickert, C.S. & Garner, B. Role of ATP-binding cassette transporters in brain lipid transport and neurological disease. J. Neurochem. 104, 1145–1166 (2008).
Kim, W.S. et al. Abca7 null mice retain normal macrophage phosphatidylcholine and cholesterol efflux activity despite alterations in adipose mass and serum cholesterol levels. J. Biol. Chem. 280, 3989–3995 (2005).
Kim, W.S., Guillemin, G.J., Glaros, E.N., Lim, C.K. & Garner, B. Quantitation of ATP-binding cassette subfamily-A transporter gene expression in primary human brain cells. Neuroreport 17, 891–896 (2006).
Jehle, A.W. et al. ATP-binding cassette transporter A7 enhances phagocytosis of apoptotic cells and associated ERK signaling in macrophages. J. Cell Biol. 174, 547–556 (2006).
Liang, Y., Buckley, T.R., Tu, L., Langdon, S.D. & Tedder, T.F. Structural organization of the human MS4A gene cluster on chromosome 11q12. Immunogenetics 53, 357–368 (2001).
Kinet, J.P. et al. Isolation and characterization of cDNAs coding for the beta subunit of the high-affinity receptor for immunoglobulin E. Proc. Natl. Acad. Sci. USA 85, 6483–6487 (1988).
Crocker, P.R., Paulson, J.C. & Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 (2007).
Tateno, H. et al. Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell signaling and innate immunity. Mol. Cell. Biol. 27, 5699–5710 (2007).
Dustin, M.L. et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell 94, 667–677 (1998).
Lynch, D.K. et al. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 278, 21805–21813 (2003).
Martinez, A. & Soriano, E. Functions of ephrin/Eph interactions in the development of the nervous system: emphasis on the hippocampal system. Brain Res. Brain Res. Rev. 49, 211–226 (2005).
Lai, K.O. & Ip, N.Y. Synapse development and plasticity: roles of ephrin/Eph receptor signaling. Curr. Opin. Neurobiol. 19, 275–283 (2009).
Coulthard, M.G. et al. Characterization of the Epha1 receptor tyrosine kinase: expression in epithelial tissues. Growth Factors 18, 303–317 (2001).
Yamazaki, T. et al. EphA1 interacts with integrin-linked kinase and regulates cell morphology and motility. J. Cell Sci. 122, 243–255 (2009).
Duffy, S.L. et al. Generation and characterization of EphA1 receptor tyrosine kinase reporter knockout mice. Genesis 46, 553–561 (2008).
Ivanov, A.I. & Romanovsky, A.A. Putative dual role of ephrin-Eph receptor interactions in inflammation. IUBMB Life 58, 389–394 (2006).
McKhann, G. et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 34, 939–944 (1984).
Mirra, S.S. et al. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology 41, 479–486 (1991).
Myers, A.J. et al. A survey of genetic human cortical gene expression. Nat. Genet. 39, 1494–1499 (2007).
Acknowledgements
For complete acknowledgments, please see the Supplementary Note. We thank the individuals and families who took part in this research and those who funded the groups who contributed to this study: Wellcome Trust; Medical Research Council (UK); Alzheimer's Research UK; the Welsh Assembly Government; Mercer's Institute for Research on Ageing; Alzheimer's Society; Ulster Garden Villages; Northern Ireland R&D Office; Royal College of Physicians; Dunhill Medical Trust; Bristol Research into Alzheimer's and Care of the Eldery; US National Institutes of Health, the Barnes Jewish Foundation; Charles and Joanne Knight Alzheimer's Research Initiative; University College London (UCL) Hospital/UCL Biomedical Centre; Lundbeck; German Federal Ministry of Education and Research Competence Network Dementia and Competence Network Degenerative Dementia; Alfried Krupp von Bohlen und Halbach-Stiftung; Intramural Research Program of the National Institute of Ageing Department of Health and Human Services; University of Antwerp, Fund for Scientific Research-Flanders; Foundation for Alzheimer Research; Methusalem Excellence grant; Federal Science Policy Office Interuniversity Attraction Poles program; Mayo AD Research Center; National Institute of Neurological Disorders and Stroke; Robert and Clarice Smith Postdoctoral Fellowship and AD Research Program; Palumbo Professorship in AD Research; Carl Edward and Susan Bass Bolch Gift; Institut Pasteur de Lille; Centre National de Génotypage; Fondation pour la Recherche Médicale Caisse; Nationale Maladie des Travailleurs Salariés, Direction Générale de la Sant; Institut de la Longévité; Agence Française de Sécurité Sanitaire des Produits de Santé; Aquitaine and Bourgogne Regional Councils; Fondation de France; French Ministry of Research/Institute national de la santé et de la recherché medicale; Eisai; Health Research Council of the Academy of Finland; Nordic Centre of Excellence in Neurodegeneration; Italian Ministry of Research; Carimonte Foundation; Italian Ministry of Health; Fondazione Monzino; Ministerio de Educación y Ciencia the Ministerio de Sanidad y Consumo; Fundación Ramón Areces; National Institute of Biomedical Imaging Bioengineering; Abbott; AstraZeneca, Bayer Schering Pharma; Bristol-Myers Squibb; Elan; Genentech; GE; GlaxoSmithKline; Innogenetics; Johnson and Johnson; Eli Lilly; Medpace; Merck; Novartis; Pfizer; Hoffman-La Roche; Schering-Plough; Synarc; Wyeth; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; US Food and Drug Administration; Foundation for the National Institutes of Health Northern California Institute for Research and Education; Dana Foundation; German National Genome Research Network; German Ministry for Education and Research; National Institute on Deafness and Other Communication Disorders; Hjartavernd; Althingi; National Heart Lung and Blood Institute; National Institute of Diabetes and Digestive and Kidney Diseases; Robert Dawson Evans Endowment; Netherlands Organisation of Scientific Research; Netherlands Genomics Initiative; Erasmus Medical Center; Netherlands organization for scientific research; Netherlands Organization for the Health Research and Development; the Research Institute for Diseases in the Elderly; Ministry of Education, Culture and Science; Ministry for Health, Welfare and Sports; European Commission and the Municipality of Rotterdam. G.W. was partly funded by the NIHR Biomedical Research Centre Programme, Oxford. The Leeds sample collection was funded by the Health Foundation.
Author information
Authors and Affiliations
Consortia
Contributions
J. Williams directed this study, assisted by M.J.O. and M.O.D. and was also helped by P.H., R.S., A.G., R.A., L.J. and D. Harold. J. Williams, P.H. and D.H. took primary responsibility for drafting the manuscript, assisted by R.S., A.G., R.A., M.O.D. and M.J.O. All authors contributed to the sample collection, sample preparation, genotyping and/or conduct of the GWAS upon which this study is based. J. Williams, R.A., P.H., R.S., A.G., C.W., J. Chapman, K.D., N.J., A.S., C. Thomas, S. Lovestone, J.P., P. Proitsi, M.K.L., C. Brayne, D.C.R., M.G., B.L., A.L., K. Morgan, K.S.B., P.A.P., D. Craig, B.M., S.T., C.H., D.M., A.D.S., S. Love, P.G.K., J.H., S. Mead, N.C.F., M. Rossor, J. Collinge, W.M., F.J., B.S., E.R., R.H., H.K., H.v.d.B., I.H., J.K., J. Wiltfang, M. Dichgans, L.F., H.H., M. Hüll, J.G., A.M.G., D.R., I.G., J.S.K.K., C.C., P.N., J.C.M., K. Mayo, K. Sleegers, K.B., S.E., P.P.D., C.V.B., G.L., N.J.B., H.G., A.M., M.T., T.W.M., M.M.N., S. Moebus, K.-H.J., N.K. and H.-E.W. contributed to clinical sample collection, ascertainment, diagnosis and preparation of samples from the independent GERAD2 sample genotyped as part of this study. R.S., D. Harold, A.G., D.R. and I.G. were responsible for procedures related to genotyping the GERAD2 sample. V.C., B.G.-B., M. Hiltunen, O.C., D.Z., M. Delepine, M.J.B., F. Pasquier, I.M., A.F.-G., E.P., O.H., E. Coto, V.A., P. Bosco, G.S., M. Mancuso, F. Panza, B.N., S. Sorbi, P. Bossù, P. Piccardi, B.A., G.A., D.S., E.S., D.G., A.B., D. Hannequin, F.L., H. Soininen, J.-C.L. and P.A. were responsible for sample collection, sample preparation, genotyping and analysis of the EADI2 Sample. S.S., A.L.D., O.L. and L.J.L., as well as M.A.I., C.M.v.D. and M.M.B.B. contributed clinical and genotypic data to the CHARGE GWAS. J.-C.L. and P.A. contributed clinical and genotypic data. M.M.C. played a leading role, along with H.B., D.W., G.W., N.M.H., E.R.L.C.V., S.B.S., J.O.A., M.B., Z.K.W., D.W.D., N.R.G.R., R.C.P., K. Morgan and S.G.Y. in sample collection, sample preparation, genotyping and analysis of the Mayo2 sample. M. Riemenschneider, T.M.F., P.F., C.R., M.K., S. Schreiber, M. Mayhaus, S.N. and S.W. were responsible for sample collection, conduct and analysis of the AD-IG GWAS. S. Steinberg, T.J., H. Stefansson, K. Stefansson, J.S., S.B. and P.V.J were responsible for sample collection, conduct and analysis of the deCODE GWAS. D. Harold and P.H. completed statistical quality control and produced association statistics under the supervision of J. Williams and P.A.H. All authors discussed the results and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have applied for an International PCT (Patent Co-operation Treaty) patent filing (European Office).
Additional information
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (see URLs). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. See URLs for a complete listing of the ADNI investigators.
Supplementary information
Supplementary Text and Figures
Supplementary Note and Supplementary Tables 1–5 (PDF 526 kb)
Supplementary Table 6
Results for SNPs at the CD2AP, EPHA1, ARID5B and CD33 loci. (XLS 93 kb)
Supplementary Table 7
Sample size tested for each SNP (XLS 75 kb)
Supplementary Table 8
Summary statistics for each dataset (XLS 285 kb)
Rights and permissions
About this article
Cite this article
Hollingworth, P., Harold, D., Sims, R. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 43, 429–435 (2011). https://doi.org/10.1038/ng.803
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.803
This article is cited by
-
CD20/MS4A1 is a mammalian olfactory receptor expressed in a subset of olfactory sensory neurons that mediates innate avoidance of predators
Nature Communications (2024)
-
Reenacting Neuroectodermal Exposure of Hematopoietic Progenitors Enables Scalable Production of Cryopreservable iPSC-Derived Human Microglia
Stem Cell Reviews and Reports (2023)
-
APOE4 impairs the microglial response in Alzheimer’s disease by inducing TGFβ-mediated checkpoints
Nature Immunology (2023)
-
Identifying Alzheimer’s genes via brain transcriptome mapping
BMC Medical Genomics (2022)
-
Exploiting family history in aggregation unit-based genetic association tests
European Journal of Human Genetics (2022)