Abstract
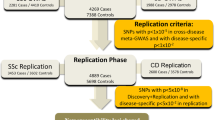
Systemic sclerosis (SSc) is an autoimmune disease characterized by fibrosis of the skin and internal organs that leads to profound disability and premature death. To identify new SSc susceptibility loci, we conducted the first genome-wide association study in a population of European ancestry including a total of 2,296 individuals with SSc and 5,171 controls. Analysis of 279,621 autosomal SNPs followed by replication testing in an independent case-control set of European ancestry (2,753 individuals with SSc (cases) and 4,569 controls) identified a new susceptibility locus for systemic sclerosis at CD247 (1q22–23, rs2056626, P = 2.09 × 10−7 in the discovery samples, P = 3.39 × 10−9 in the combined analysis). Additionally, we confirm and firmly establish the role of the MHC (P = 2.31 × 10−18), IRF5 (P = 1.86 × 10−13) and STAT4 (P = 3.37 × 10−9) gene regions as SSc genetic risk factors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
16 April 2010
In the version of this article initially published online, the name of author Annemie J. Schuerwegh was misspelled. The error has been corrected for the print, PDF, and HTML versions of this article.
23 March 2011
In the version of this article initially published, incorrect affiliations were published for Lorenzo Beretta and Raffaella Scorza. The correct affiliation for Lorenzo Beretta and Raffaella Scorza is "Referral Center for Systemic Autoimmune Diseases, Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico and University of Milan". The error has been corrected in the HTML and PDF versions of the article.
References
Gabrielli, A., Avvedimento, E.V. & Krieg, T. Scleroderma. N. Engl. J. Med. 360, 1989–2003 (2009).
Jimenez, S.A. & Derk, C.T. Following the molecular pathways toward an understanding of the pathogenesis of systemic sclerosis. Ann. Intern. Med. 140, 37–50 (2004).
Agarwal, S.K., Tan, F.K. & Arnett, F.C. Genetics and genomic studies in scleroderma (systemic sclerosis). Rheum. Dis. Clin. North Am. 34, 17–40 (2008).
Arnett, F.C. et al. Major Histocompatibility Complex (MHC) class II alleles, haplotypes, and epitopes which confer susceptibility or protection in the fibrosing autoimmune disease systemic sclerosis: analyses in 1300 Caucasian, African-American and Hispanic cases and 1000 controls. Ann. Rheum. Dis. published online (12 July 2009).
Rueda, B. et al. The STAT4 gene influences the genetic predisposition to systemic sclerosis phenotype. Hum. Mol. Genet. 18, 2071–2077 (2009).
Tsuchiya, N. et al. Association of STAT4 polymorphism with systemic sclerosis in a Japanese population. Ann. Rheum. Dis. 68, 1375–1376 (2009).
Ito, I. et al. Association of a functional polymorphism in the IRF5 region with systemic sclerosis in a Japanese population. Arthritis Rheum. 60, 1845–1850 (2009).
Dieudé, P. et al. Association between the IRF5 rs2004640 functional polymorphism and systemic sclerosis: a new perspective for pulmonary fibrosis. Arthritis Rheum. 60, 225–233 (2009).
Gregersen, P.K. & Olsson, L.M. Recent advances in the genetics of autoimmune disease. Annu. Rev. Immunol. 27, 363–391 (2009).
Hunter, D.J. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874 (2007).
Yeager, M. et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39, 645–649 (2007).
Stefansson, H. et al. Large recurrent microdeletions associated with schizophrenia. Nature 455, 232–236 (2008).
Krawczak, M. et al. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 9, 55–61 (2006).
Wellcome Trust Case-Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Call, M.E. & Wucherpfennig, K.W. Molecular mechanisms for the assembly of the T cell receptor-CD3 complex. Mol. Immunol. 40, 1295–1305 (2004).
Krishnan, S. et al. Increased caspase-3 expression and activity contribute to reduced CD3zeta expression in systemic lupus erythematosus T cells. J. Immunol. 175, 3417–3423 (2005).
Krishnan, S. et al. Generation and biochemical analysis of human effector CD4 T cells: alterations in tyrosine phosphorylation and loss of CD3ζ expression. Blood 97, 3851–3859 (2001).
Krishnan, S., Warke, V.G., Nambiar, M.P., Tsokos, G.C. & Farber, D.L. The FcR gamma subunit and Syk kinase replace the CD3 zeta-chain and ZAP-70 kinase in the TCR signaling complex of human effector CD4 T cells. J. Immunol. 170, 4189–4195 (2003).
Gorman, C.L. et al. Polymorphisms in the CD3Z gene influence TCRzeta expression in systemic lupus erythematosus patients and healthy controls. J. Immunol. 180, 1060–1070 (2008).
Warchol, T. et al. The CD3Z 844 T>A polymorphism within the 3′-UTR of CD3Z confers increased risk of incidence of systemic lupus erythematosus. Tissue Antigens 74, 68–72 (2009).
Gourh, P. et al. Association of the PTPN22 R620W polymorphism with anti-topoisomerase I- and anticentromere antibody-positive systemic sclerosis. Arthritis Rheum. 54, 3945–3953 (2006).
Anonymous. Preliminary criteria for the classification of systemic sclerosis (scleroderma). Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Arthritis Rheum. 23, 581–590 (1980).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Tian, C. et al. European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Mol. Med. 15, 371–383 (2009).
Acknowledgements
This work was supported by the following grants: T.R.D.J.R. was funded by the VIDI laureate from the Dutch Association of Research (NWO) and Dutch Arthritis Foundation (National Reumafonds). J.M. was funded by GEN-FER from the Spanish Society of Rheumatology, SAF2009-11110 from the Spanish Ministry of Science, CTS-4977 from Junta de Andalucía, Spain and in part by Redes Temáticas de Investigación Cooperativa Sanitaria Program, RD08/0075 (RIER) from Instituto de Salud Carlos III (ISCIII), Spain (J.M.). R.B. is supported by the I3P Consejo Superior de Investigaciones Científicas program funded by the 'Fondo Social Europeo'. B.Z.A. is supported by the Netherlands Organization for Health Research and Development (ZonMW grant 016.096.121). B.K. is supported by the Dutch Diabetes Research Foundation (grant 2008.40.001) and the Dutch Arthritis Foundation (Reumafonds, grant NR 09-1-408). Genotyping of the Dutch control samples was sponsored by US National Insitutes of Mental Health funding, R01 MH078075 (R.O.A.). The German controls were from the PopGen biobank (to B.K.). The PopGen project received infrastructure support through the German Research Foundation excellence cluster 'Inflammation at Interfaces'. The US analyses were supported by the US National Institutes of Health and National Institute of Arthritis and Musculoskeletal Diseases (NIH-NIAMS) R01 AR055258, Two-Stage Genome Wide Association Study in Systemic Sclerosis, (M.D.M.) and by the NIH-NIAMS Center of Research Translation (CORT) in SSc (P50AR054144) (F.C.A.), the NIH-NIAMS SSc Family Registry and DNA Repository (N01-AR-0-2251) (M.D.M.), University of Texas Health Science Center-Houston Center for Clinical and Translational Sciences (Houston Clinical and Translational Science Awards Program) (NIH-National Center for Research Resources 3UL1RR024148) (F.C.A.), NIH-NIAMS K08 Award (K08AR054404) (S.K.A.), SSc Foundation New Investigator Award (S.K.A.).
Author information
Authors and Affiliations
Consortia
Contributions
Study Design: T.R.D.J.R., O.G., B.R., J.-E.M., B.P.C.K., F.C.A., J.M., M.D.M.
Collection of data: T.R.D.J.R., M.J.C., M.C.V., A.E.V., A.J.S., J.C.B., B.A.L., A.-M.H.-V., R.A.O., G.R., N.H., C.P.S., N.O.-C., M.A.G.-G., M.F.G.-E., P.A., J.v.L., A.H., J.W., R.H., V.S., F.d.K., F.H., M.M.C., R.M., P.S., R.W., A.K., H.K., E.d.B., T.W., L.P., L.K., L.B., R.S., J.V., M.H., P.G., J.L.N., F.M.W., L.H., P.C., S.A.
Interpretation and analysis of results: T.R.D.J.R., O.G., B.R., J.-E.M., B.Z.A., R.P.-M., J.Y., Y.H., S.-F.W., R.v.'t.S., P.G., A.T.L., C.I.A., S.K.A., B.P.C.K., J.M., M.D.M., A.I., P.C., S.A., P.K.G.
Critical reading of manuscript: T.R.D.J.R., O.G., B.R., J.-E.M., B.Z.A., J.Y., M.J.C., M.C.V., A.E.V., A.J.S., J.C.B., P.L.C.M.v.R., R.v.S., B.A.L., A.-M.H.-V., G.R., N.H., C.P.S., N.O.-C., M.A.G.-G., M.F.G.-E., P.A., J.v.L., A.H., J.W., R.H., V.S., F.d.K., F.H., M.M.C., R.M., P.S., R.W., A.K., H.K., E.d.B., T.W., L.P., L.B., R.S., J.V., M.H., P.G., C.I.A., J.L.N., F.M.W., L.H., S.K.A., P.G., F.K.T., B.P.C.K., F.C.A., J.M., M.D.M., P.K.G.
Project conception: T.R.D.J.R., B.P.C.K., F.C.A., J.M., M.D.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–5, Supplementary Figures 1–4 and Supplementary Note (PDF 2108 kb)
Rights and permissions
About this article
Cite this article
Radstake, T., Gorlova, O., Rueda, B. et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet 42, 426–429 (2010). https://doi.org/10.1038/ng.565
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.565
This article is cited by
-
Broadening our understanding of genetic risk for scleroderma/systemic sclerosis by querying the chromatin architecture surrounding the risk haplotypes
BMC Medical Genomics (2021)
-
Updates on genetics in systemic sclerosis
Inflammation and Regeneration (2021)
-
Esophageal dysmotility and lung disease in patients with systemic sclerosis: is there a possible association or correlation? A retrospective chart review
Rheumatology International (2021)
-
Dysregulation of Type I Interferon Signaling in Systemic Sclerosis: a Promising Therapeutic Target?
Current Treatment Options in Rheumatology (2021)
-
Epigenomics and transcriptomics of systemic sclerosis CD4+ T cells reveal long-range dysregulation of key inflammatory pathways mediated by disease-associated susceptibility loci
Genome Medicine (2020)