Abstract
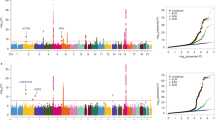
Lung cancer is the most common cause of death from cancer worldwide, and its incidence is increasing in East Asian and Western countries. To identify genetic factors that modify the risk of lung adenocarcinoma, we conducted a genome-wide association study in a Japanese cohort, with replication in two independent studies in Japanese and Korean individuals, in a total of 2,098 lung adenocarcinoma cases and 11,048 controls. The combined analyses identified two susceptibility loci for lung adenocarcinoma: TERT (rs2736100, combined P = 2.91 × 10−11, odds ratio (OR) = 1.27) and TP63 (rs10937405, combined P = 7.26 × 10−12, OR = 1.31). Fine mapping of the region containing TP63 showed that a SNP (rs4488809) in intron 1 of TP63 showed the most significant association. Our results suggest that genetic variation in TP63 may influence susceptibility to lung adenocarcinoma in East Asian populations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Jemal, A. et al. Cancer statistics, 2009. CA Cancer J. Clin. 59, 225–249 (2009).
Parkin, D.M., Bray, F., Ferlay, J. & Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 55, 74–108 (2005).
Daigo, Y. & Nakamura, Y. From cancer genomics to thoracic oncology: discovery of new biomarkers and therapeutic targets for lung and esophageal carcinoma. Gen. Thorac. Cardiovasc. Surg. 56, 43–53 (2008).
Toyoda, Y., Nakayama, T., Ioka, A. & Tsukuma, H. Trends in lung cancer incidence by histological type in Osaka, Japan. Jpn. J. Clin. Oncol. 38, 534–539 (2008).
Sobue, T. et al. Trend of lung cancer incidence rate by histological type: a population-based study in Osaka, Japan. Jpn. J. Cancer Res. 90, 6–15 (1999).
Thun, M.J. et al. Cigarette smoking and changes in the histopathology of lung cancer. J. Natl. Cancer Inst. 89, 1580–1586 (1997).
Devesa, S.S., Bray, F., Vizcaino, A.P. & Parkin, D.M. International lung cancer trends by histological type. Int. J. Cancer 117, 294–299 (2005).
Janssen-Heijnen, M.L. & Coebergh, J.W. The changing epidemiology of lung cancer in Europe. Lung Cancer 41, 245–258 (2003).
Yang, C.H. EGFR tyrosine kinase inhibitors for the treatment of NSCLC in East Asia: present and future. Lung Cancer 60 (Suppl. 2), S23–S30 (2008).
Jee, S.H., Kim, I.S., Suh, I., Shin, D. & Appel, L.J. Projected mortality from lung cancer in South Korea, 1980–2004. Int. J. Epidemiol. 27, 365–369 (1998).
Liam, C.K., Pang, Y.K., Leow, C.H., Poosparajah, S. & Menon, A. Changes in the distribution of lung cancer cell types and patient demography in a developing multiracial Asian country: experience of a university teaching hospital. Lung Cancer 53, 23–30 (2006).
Fukuoka, M. et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 21, 2237–2246 (2003).
Huang, S.F. et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin. Cancer Res. 10, 8195–8203 (2004).
Marchetti, A. et al. EGFR mutations in non-small-cell lung cancer. J. Clin. Oncol. 23, 857–865 (2005).
Matakidou, A., Eisen, T. & Houlston, R.S. Systematic review of the relationship between family history and lung cancer risk. Br. J. Cancer 93, 825–833 (2005).
Zhang, Y. et al. Family history of cancer and risk of lung cancer among nonsmoking Chinese women. Cancer Epidemiol. Biomarkers Prev. 16, 2432–2435 (2007).
Hung, R.J. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 452, 633–637 (2008).
Amos, C.I. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 40, 616–622 (2008).
Thorgeirsson, T.E. et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 452, 638–642 (2008).
McKay, J.D. et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 40, 1404–1406 (2008).
Wang, Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 40, 1407–1409 (2008).
Broderick, P. et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 69, 6633–6641 (2009).
Landi, M.T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 85, 679–691 (2009).
Wu, C. et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 69, 5065–5072 (2009).
Moll, U.M. & Slade, N. p63 and p73: roles in development and tumor formation. Mol. Cancer Res. 2, 371–386 (2004).
Flores, E.R. The roles of p63 in cancer. Cell Cycle 6, 300–304 (2007).
Katoh, I., Aisaki, K., Kurata, S., Ikawa, S. & Ikawa, Y. p51A (TAp63gamma), a p53 homolog, accumulates in response to DNA damage for cell regulation. Oncogene 19, 3126–3130 (2000).
Petitjean, A. et al. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis 29, 273–281 (2008).
Kiemeney, L.A. et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 40, 1307–1312 (2008).
Nakamura, Y. The BioBank Japan project. Clin. Adv. Hematol. Oncol. 5, 696–697 (2007).
Ohnishi, Y. et al. A high-throughput SNP typing system for genome-wide association studies. J. Hum. Genet. 46, 471–477 (2001).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Barrett, J.C., Fry, B., Maller, J. & Daly, M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Acknowledgements
We thank the staff of the Laboratory for Genotyping Development, Center for Genomic Medicine, RIKEN and the Human Genome Center, Institute of Medical Science, The University of Tokyo for their contribution to SNP genotyping. We also thank members of the BioBank Japan project, the Rotary Club of Osaka-Midosuji District 2660 Rotary International in Japan, and Research Institute and Hospital, National Cancer Center, Korea for supporting our study. Y.D. is a member of the Shiga Cancer Treatment Project supported by Shiga Prefecture (Japan). This work was conducted as a part of the BioBank Japan Project and supported by the Ministry of Education, Culture, Sports, Sciences and Technology of the Japanese government. Management of second replication samples in Korea was supported by grants 0710221 and 0940620 from the National Cancer Center, Korea.
Author information
Authors and Affiliations
Contributions
Y.N. conceived the study; D.M., M.K., Y.N. and Y.D. designed the study; D.M., N.H., M.K. and Y.D. performed genotyping; D.M., M.K., Y.N. and Y.D. wrote the manuscript; A.T., T.M., T. Tsunoda and N.K. performed data analysis at the genome-wide phase; Y.N. and M.K. managed DNA samples belong to BioBank Japan; K.-A.Y., J.K., G.-K.L., J.I.Z. and J.S.L. managed second replication samples in Korea; D.M. and Y.D. summarized the results; Y.N., T. Takahashi, K.C., J.I. and Y.D. obtained funding for the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 and Supplementary Tables 1–6 (PDF 783 kb)
Rights and permissions
About this article
Cite this article
Miki, D., Kubo, M., Takahashi, A. et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet 42, 893–896 (2010). https://doi.org/10.1038/ng.667
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.667
This article is cited by
-
Contribution of an Asian-prevalent HLA haplotype to the risk of HBV-related hepatocellular carcinoma
Scientific Reports (2023)
-
Ethnicity-specific association between TERT rs2736100 (A > C) polymorphism and lung cancer risk: a comprehensive meta-analysis
Scientific Reports (2023)
-
Genome-wide association study of lung adenocarcinoma in East Asia and comparison with a European population
Nature Communications (2023)
-
Interleukin-17A mediates tobacco smoke–induced lung cancer epithelial-mesenchymal transition through transcriptional regulation of ΔNp63α on miR-19
Cell Biology and Toxicology (2022)
-
Susceptibility loci for pancreatic cancer in the Brazilian population
BMC Medical Genomics (2021)