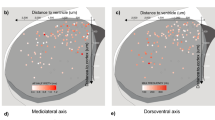
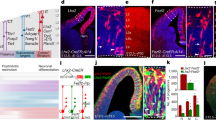
Abstract
The extent to which individual neurons are interconnected selectively within brain circuits is an unresolved problem in neuroscience. Neurons can be organized into preferentially interconnected microcircuits, but whether this reflects genetically defined subpopulations is unclear. We found that the principal neurons in the main subdivisions of the hippocampus consist of distinct subpopulations that are generated during distinct time windows and that interconnect selectively across subdivisions. In two mouse lines in which transgene expression was driven by the neuron-specific Thy1 promoter, transgene expression allowed us to visualize distinct populations of principal neurons with unique and matched patterns of gene expression, shared distinct neurogenesis and synaptogenesis time windows, and selective connectivity at dentate gyrus-CA3 and CA3-CA1 synapses. Matched subpopulation marker genes and neuronal subtype markers mapped near clusters of olfactory receptor genes. The nonoverlapping matched timings of synaptogenesis accounted for the selective connectivities of these neurons in CA3. Therefore, the hippocampus contains parallel connectivity channels assembled from distinct principal neuron subpopulations through matched schedules of synaptogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Brown, S.P. & Hestrin, S. Cell-type identity: a key to unlocking the function of neocortical circuits. Curr. Opin. Neurobiol. 19, 415–421 (2009).
Yoshimura, Y. & Callaway, E.M. Fine-scale specificity of cortical networks depends on inhibitory cell type and connectivity. Nat. Neurosci. 8, 1552–1559 (2005).
Song, S., Sjostrom, P.J., Reigl, M., Nelson, S. & Chklovskii, D.B. Highly nonramdom features of synaptic connectivity in local cortical circuits. PLoS Biol. 3, e68 (2005).
Kampa, B.M., Letzkus, J.J. & Stuart, G.J. Cortical feed-forward networks for binding different streams of sensory information. Nat. Neurosci. 9, 1472–1473 (2006).
Brown, S.P. & Hestrin, S. Intracortical circuits of pyramidal neurons reflect their long-range axonal targets. Nature 457, 1133–1136 (2009).
Petreanu, L., Mao, T., Sternson, S.M. & Svoboda, K. The subcellular organization of neocortical excitatory connections. Nature 457, 1142–1145 (2009).
Yu, Y.C., Bultje, R.S., Wang, X. & Shi, S.H. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature 458, 501–504 (2009).
Caroni, P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. J. Neurosci. Methods 71, 3–9 (1997).
Feng, G. et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 (2000).
De Paola, V., Arber, S. & Caroni, P. AMPA receptors regulate dynamic equilibrium of presynaptic terminals in mature hippocampal networks. Nat. Neurosci. 6, 491–500 (2003).
Haverkamp, S. et al. The primordial, blue-cone color system of the mouse retina. J. Neurosci. 25, 5438–5445 (2005).
Amaral, D. & Lavenex, P. Hippocampal neuroanatomy. in The Hippocampus Book (eds. Andersen, P. et al.) 37–110 (Oxford Univ. Press, 2007).
Lein, E.S., Zhao, X. & Gage, F.H. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J. Neurosci. 24, 3879–3889 (2004).
Thompson, C.L. et al. Genomic anatomy of the hippocampus. Neuron 60, 1010–1021 (2008).
Kamme, F. et al. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J. Neurosci. 23, 3607–3615 (2003).
Gogolla, N., Galimberti, I., Deguchi, Y. & Caroni, P. Wnt signaling mediates experience-related regulation of synapse numbers and mossy fiber connectivities in the adult hippocampus. Neuron 62, 510–525 (2009).
Galimberti, I., Bednarek, E., Donato, F. & Caroni, P. EphA4 signaling in juveniles establishes topographic specificity of structural plasticity in the hippocampus. Neuron 65, 627–642 (2010).
Saxena, S., Cabuy, E. & Caroni, P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 12, 627–636 (2009).
Noctor, S.C., Flint, A.C., Weissman, T.A., Dammerman, R.S. & Kriegstein, A.R. Neurons derived from radial glia cells establish radial units in neocortex. Nature 409, 714–720 (2001).
Altman, J. & Bayer, S.A. Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. J. Comp. Neurol. 301, 325–342 (1990).
Tole, S. & Grove, E.A. Detailed field pattern is intrinsic to the embryonic mouse hippocampus early in neurogenesis. J. Neurosci. 21, 1580–1589 (2001).
Chehrehasa, F., Meedeniya, A.C., Dwyer, P., Abrahamsen, G. & Mackay-Sim, A. EdU, a new thymidine analogue for labeling proliferating cells in the nervous system. J. Neurosci. Methods 177, 122–130 (2009).
Hodge, R.D. et al. Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J. Neurosci. 28, 3707–3717 (2008).
Wojtowicz, J.M. & Kee, N. BrdU assay for neurogenesis in rodents. Nat. Protoc. 1, 1399–1405 (2006).
Li, G. et al. Hilar mossy cells share developmental influences with dentate granule neurons. Dev. Neurosci. 30, 255–261 (2008).
Altman, J. & Bayer, S.A. Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J. Comp. Neurol. 301, 365–381 (1990).
Godfrey, P.A., Malnic, B. & Buck, L.B. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. USA 101, 2156–2161 (2004).
Dijk, F., Leeuwen, S.v. & Kamphuis, W. Differential effects of ischemia/reperfusion on amacrine cell subtype–specific transcript levels in the rat retina. Brain Res. 1026, 194–204 (2004).
Zylka, M.J., Rice, F.L. & Anderson, D.J. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron 45, 17–25 (2005).
Jones, S.P., Rahimi, O., O'Boyle, M.P., Diaz, D.L. & Claiborne, B.J. Maturation of granule cell dendrites after mossy fiber arrival in hippocampal field CA3. Hippocampus 13, 413–427 (2003).
Tremblay, M.E., Riad, M., Chierzi, S., Murai, K.K. & Pasquale, E. Developmental course of EphA4 cellular and subcellular localization in the postnatal rat hippocampus. J. Comp. Neurol. 512, 798–813 (2009).
Frotscher, M., Zhao, S. & Förster, E. Development of cell and fiber layers in the dentate gyrus. Prog. Brain Res. 163, 133–142 (2007).
Isshiki, T., Pearson, B., Holbrook, S. & Doe, C.Q. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521 (2001).
Noctor, S.C., Martínez-Cerdeño, V., Ivic, L. & Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004).
Baumgardt, M., Karlsson, D., Terriente, J., Diaz-Benjumea, F.J. & Thor, S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell 139, 969–982 (2009).
Petrovic, M. & Hummel, T. Temporal identity in axonal target layer recognition. Nature 456, 800–803 (2008).
Baek, M. & Mann, R.S. Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosphila. J. Neurosci. 29, 6904–6916 (2009).
Elliott, J., Jolicoeur, C., Ramamurthy, V. & Cayouette, M. Ikaros confers early temporal competence to mouse retinal progenitor cell. Neuron 60, 26–39 (2008).
Gaspard, N. et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455, 351–357 (2008).
Miyoshi, G., Butt, S.J.B., Takebayashi, H. & Fishell, G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J. Neurosci. 27, 7786–7798 (2007).
Gelman, D.M. & Marin, O. Generation of interneuron diversity in the mouse cerebral cortex. Eur. J. Neurosci. 31, 2136–2141 (2010).
Kamme, F. et al. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J. Neurosci. 23, 3607–3615 (2003).
Jefferis, G.S., Marin, E.C., Stocker, R.F. & Luo, L. Target neuron prespecification in the olfactory map of Drosophila. Nature 414, 204–208 (2001).
McLean, D.L. & Fetcho, J.R. Spinal interneurons differentiate sequentially from those driving the fastest swimming movements in larval zebrafish to those driving the slowest ones. J. Neurosci. 29, 13566–13577 (2009).
Espinosa, J.S. & Luo, L. Timing neurogenesis and differentiation: insights from quantitative clonal analyses of cerebellar granule cells. J. Neurosci. 28, 2301–2312 (2008).
Toni, N. et al. Neurons born in the adult dentate gyrus form functional synapses with target cells. Nat. Neurosci. 11, 901–907 (2008).
Colgin, L.L., Moser, E.I. & Moser, M.B. Understanding memory through hippocampal remapping. Trends Neurosci. 31, 469–477 (2008).
Pinaud, R. et al. Detection of two mRNA species at single-cell resolution by double-fluorescence in situ hybridization. Nat. Protoc. 3, 1370–1379 (2008).
Knott, G.W., Holtmaat, A., Trachtenberg, J.T., Svoboda, K. & Welker, E. A protocol for preparing GFP-labeled neurons previously imaged in vivo and in slice preparations for light and electron microscopic analysis. Nat. Protoc. 4, 1145–1156 (2009).
Gogolla, N., Galimberti, I., DePaola, V. & Caroni, P. Preparation of organotypic slice cultures for long term live imaging. Nat. Protoc. 1, 1223–1226 (2006).
Acknowledgements
We thank S. Arber, A. Lüthi and B. Roska (Friedrich Miescher Institute; FMI) for comments on the manuscript, C. Genoud (FMI) for assistance with immuno-electron microscopy, M. Wiechert and A. Ponti (FMI) for data analysis assistance, and S. Arber for the rabies-mCherry virus. The FMI is part of the Novartis Research Foundation.
Author information
Authors and Affiliations
Contributions
Y.D. conceived and carried out the gene expression and the adult subpopulation mapping analysis. F.D. conceived and carried out the neurogenesis and synaptogenesis analysis and parts of the electron microscopy and connectivity analysis. I.G. conceived and carried out most of the connectivity analysis. E.C. carried out and optimized the cell genomics experiments. P.C. helped devise the experiments and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12 (PDF 3728 kb)
Rights and permissions
About this article
Cite this article
Deguchi, Y., Donato, F., Galimberti, I. et al. Temporally matched subpopulations of selectively interconnected principal neurons in the hippocampus. Nat Neurosci 14, 495–504 (2011). https://doi.org/10.1038/nn.2768
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2768
This article is cited by
-
Formation of memory assemblies through the DNA-sensing TLR9 pathway
Nature (2024)
-
Neurogenic timing of the inferior olive subdivisions is related to the olivocerebellar projection topography
Scientific Reports (2023)
-
Associations between in vitro, in vivo and in silico cell classes in mouse primary visual cortex
Nature Communications (2023)
-
Image cutting in video media technology application based on detection algorithm
International Journal of System Assurance Engineering and Management (2023)
-
Preconfigured dynamics in the hippocampus are guided by embryonic birthdate and rate of neurogenesis
Nature Neuroscience (2022)