Abstract
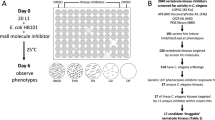
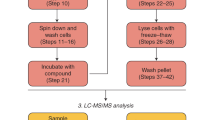
The resistance of Caenorhabditis elegans to pharmacological perturbation limits its use as a screening tool for novel small bioactive molecules. One strategy to improve the hit rate of small-molecule screens is to preselect molecules that have an increased likelihood of reaching their target in the worm. To learn which structures evade the worm's defenses, we performed the first survey of the accumulation and metabolism of over 1,000 commercially available drug-like small molecules in the worm. We discovered that fewer than 10% of these molecules accumulate to concentrations greater than 50% of that present in the worm's environment. Using our dataset, we developed a structure-based accumulation model that identifies compounds with an increased likelihood of bioavailability and bioactivity, and we describe structural features that facilitate small-molecule accumulation in the worm. Preselecting molecules that are more likely to reach a target by first applying our model to the tens of millions of commercially available compounds will undoubtedly increase the success of future small-molecule screens with C. elegans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Burns, A.R. et al. High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nat. Protoc. 1, 1906–1914 (2006).
Kwok, T.C.Y. et al. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature 441, 91–95 (2006).
Petrascheck, M., Ye, X. & Buck, L.B. An antidepressant that extends lifespan in adult Caenorhabditis elegans. Nature 450, 553–556 (2007).
Kokel, D., Li, Y., Qin, J. & Xue, D. The nongenotoxic carcinogens naphthalene and para-dichlorobenzene suppress apoptosis in Caenorhabditis elegans. Nat. Chem. Biol. 2, 338–345 (2006).
Kwok, T.C. et al. A genetic screen for dihydropyridine (DHP)-resistant worms reveals new residues required for DHP-blockage of mammalian calcium channels. PLoS Genet. 4, e1000067 (2008).
Jones, A.K., Buckingham, S.D. & Sattelle, D.B. Chemistry-to-gene screens in Caenorhabditis elegans. Nat. Rev. Drug Discov. 4, 321–330 (2005).
Kaminsky, R. et al. A new class of anthelmintics effective against drug-resistant nematodes. Nature 452, 176–180 (2008).
Kaletta, T. & Hengartner, M.O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 5, 387–398 (2006).
Broeks, A., Janssen, H.W., Calafat, J. & Plasterk, R.H. A P-glycoprotein protects Caenorhabditis elegans against natural toxins. EMBO J. 14, 1858–1866 (1995).
Rand, J.B. & Johnson, C.D. Genetic pharmacology: interactions between drugs and gene products in Caenorhabditis elegans. in Methods in Cell Biology, 48 (eds. Epstein, H.F. & Shakes, D.C.) 187–204 (Academic, San Diego, 1995).
Choy, R.K. & Thomas, J.H. Fluoxetine-resistant mutants in C. elegans define a novel family of transmembrane proteins. Mol. Cell 4, 143–152 (1999).
Cox, G.N., Kusch, M. & Edgar, R.S. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J. Cell Biol. 90, 7–17 (1981).
Avery, L. & Shtonda, B.B. Food transport in the C. elegans pharynx. J. Exp. Biol. 206, 2441–2457 (2003).
Lindblom, T.H. & Dodd, A.K. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J. Exp. Zool. A. Comp. Exp. Biol. 305, 720–730 (2006).
Jospin, M., Jacquemond, V., Mariol, M.C., Segalat, L. & Allard, B. The L-type voltage-dependent Ca2+ channel EGL-19 controls body wall muscle function in Caenorhabditis elegans. J. Cell Biol. 159, 337–348 (2002).
Franks, C.J. et al. Ionic basis of the resting membrane potential and action potential in the pharyngeal muscle of Caenorhabditis elegans. J. Neurophysiol. 87, 954–961 (2002).
Irwin, J.J. & Shoichet, B.K. ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 (2005).
Herre, S. & Pragst, F. Shift of the high-performance liquid chromatographic retention times of metabolites in relation to the original drug on an RP8 column with acidic mobile phase. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 692, 111–126 (1997).
Herzler, M., Herre, S. & Pragst, F. Selectivity of substance identification by HPLC–DAD in toxicological analysis using a UV spectra library of 2682 compounds. J. Anal. Toxicol. 27, 233–242 (2003).
Lipinski, C.A., Lombardo, F., Dominy, B.W. & Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 (2001).
Kocisko, D.A. et al. New inhibitors of scrapie-associated prion protein formation in a library of 2000 drugs and natural products. J. Virol. 77, 10288–10294 (2003).
Eddershaw, P. & Dickins, M. Phase I metabolism. in A Handbook of Bioanalysis and Drug Metabolism (ed. Evans, G.) 208–221 (CRC Press, Boca Raton, Florida, USA, 2004).
Manchee, G., Dickins, M. & Pickup, E. Phase II enzymes. in A Handbook of Bioanalysis and Drug Metabolism (ed. Evans, G.) 222–243 (CRC Press, Boca Raton, Florida, USA, 2004).
Xia, X., Maliski, E.G., Gallant, P. & Rogers, D. Classification of kinase inhibitors using a Bayesian model. J. Med. Chem. 47, 4463–4470 (2004).
Rogers, D., Brown, R.D. & Hahn, M. Using extended-connectivity fingerprints with Laplacian-modified Bayesian analysis in high-throughput screening follow-up. J. Biomol. Screen. 10, 682–686 (2005).
Durant, J.L., Leland, B.A., Henry, D.R. & Nourse, J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 42, 1273–1280 (2002).
Kerwar, S.S. Pharmacologic properties of fenbufen. Am. J. Med. 75, 62–69 (1983).
Flower, D.R. On the properties of bit string-based measures of chemical similarity. J. Chem. Inf. Comput. Sci. 38, 379–386 (1998).
Hert, J., Irwin, J.J., Laggner, C., Keiser, M.J. & Shoichet, B.K. Quantifying biogenic bias in screening libraries. Nat. Chem. Biol. 5, 479–483 (2009).
Bemis, G.W. & Murcko, M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893 (1996).
Shelat, A.A. & Guy, R.K. Scaffold composition and biological relevance of screening libraries. Nat. Chem. Biol. 3, 442–446 (2007).
Hoon, S. et al. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 4, 498–506 (2008).
Young, D.W. et al. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat. Chem. Biol. 4, 59–68 (2008).
Horton, D.A., Bourne, G.T. & Smythe, M.L. The combinatorial synthesis of bicyclic privileged structures or privileged substructures. Chem. Rev. 103, 893–930 (2003).
Klekota, J. & Roth, F.P. Chemical substructures that enrich for biological activity. Bioinformatics 24, 2518–2525 (2008).
Evans, B.E. et al. Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J. Med. Chem. 31, 2235–2246 (1988).
Mason, J.S. et al. New 4-point pharmacophore method for molecular similarity and diversity applications: overview of the method and applications, including a novel approach to the design of combinatorial libraries containing privileged substructures. J. Med. Chem. 42, 3251–3264 (1999).
Hajduk, P.J., Bures, M., Praestgaard, J. & Fesik, S.W. Privileged molecules for protein binding identified from NMR-based screening. J. Med. Chem. 43, 3443–3447 (2000).
Chen, Y. & Shoichet, B.K. Molecular docking and ligand specificity in fragment-based inhibitor discovery. Nat. Chem. Biol. 5, 358–364 (2009).
Garzon-Aburbeh, A., Poupaert, J.H., Claesen, M. & Dumont, P. A lymphotropic prodrug of L-dopa: synthesis, pharmacological properties, and pharmacokinetic behavior of 1,3-dihexadecanoyl-2-[(S)-2-amino-3-(3,4-dihydroxyphenyl)prop anoyl]propane-1,2,3-triol. J. Med. Chem. 29, 687–691 (1986).
Inturrisi, C.E. et al. Evidence from opiate binding studies that heroin acts through its metabolites. Life Sci. 33 Suppl 1: 773–776 (1983).
Hou, B., Lim, E.K., Higgins, G.S. & Bowles, D.J. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279, 47822–47832 (2004).
Cline, M.S. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 (2007).
Acknowledgements
We thank C. Cummins for critical comments on the manuscript; V. Wong and G. Selman for technical assistance early on in the project; S. Pan at the University of California, Riverside, for MS and MS-MS analyses; and A. Young of the Advanced Instrumentation for Molecular Structure Mass Spectrometry Laboratory at the University of Toronto for accurate mass MS analyses. This work was supported by Canadian Institutes of Health Research (CIHR) operating grants to P.J.R. (grant number 68813) and G.G. and C.N. (MOP-81340), Natural Sciences and Engineering Research Council of Canada support to G.D.B., a Natural Sciences and Engineering Research Council of Canada Graduate Scholarship doctoral award to A.R.B. and a Marie Curie Fellowship to I.M.W. G.G. and P.J.R. are Canadian Research Chairs in Chemical Biology and Molecular Neurobiology, respectively.
Author information
Authors and Affiliations
Contributions
A.R.B. did the wet-lab work and analyzed the MS data with guidance from S.R.C. and P.J.R. I.M.W. did the computational analysis with guidance from A.R.B., J.W., M.T., G.D.B., G.G., C.N. and P.J.R. The project was conceived by P.J.R., S.R.C. and A.R.B., and the paper was written by A.R.B. and P.J.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–10, Supplementary Table 1 and Supplementary Methods (PDF 15463 kb)
Supplementary Data 1
An Excel file of the top-scoring 5% of the ZINC collection of commercially available small molecules as ranked by our structure-accumulation model. In the version of this supplementary file originally posted online, the file contained only 65,536 lines instead of 683,493 lines. The error has been corrected in this file as of 18 June 2010. Please unzip and open with the 2007 or a newer version of Excel. (ZIP 34824 kb)
Supplementary Data 2
The xml file of our structure-accumulation model script for use with Pipeline Pilot. (XML 77 kb)
Supplementary Data 3
An sdf file of the small-molecule training set used to build our structure-accumulation model. In the version of this supplementary file originally posted online, the file was displayed as a pdf rather than in the correct sdf file format. The error has been corrected in this file as of 18 June 2010. (SDF 749 kb)
Supplementary Data 4
A list of available open-source software that could be used to generate a similar structure-accumulation model if Pipeline Pilot is not available. (PDF 40 kb)
Rights and permissions
About this article
Cite this article
Burns, A., Wallace, I., Wildenhain, J. et al. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol 6, 549–557 (2010). https://doi.org/10.1038/nchembio.380
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.380
This article is cited by
-
Parasitic nematodes activate chemicals that can kill them
Nature (2023)
-
Unraveling effects of anti-aging drugs on C. elegans using liposomes
GeroScience (2023)
-
Selective control of parasitic nematodes using bioactivated nematicides
Nature (2023)
-
Property space mapping of Pseudomonas aeruginosa permeability to small molecules
Scientific Reports (2022)
-
Nematodes as Ghosts of Land Use Past: Elucidating the Roles of Soil Nematode Community Studies as Indicators of Soil Health and Land Management Practices
Applied Biochemistry and Biotechnology (2022)