Abstract
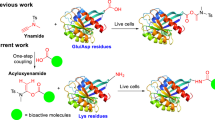
Targeted covalent inhibition of disease-associated proteins has become a powerful methodology in the field of drug discovery, leading to the approval of new therapeutics. Nevertheless, current approaches are often limited owing to their reliance on a cysteine residue to generate the covalent linkage. Here we used aryl boronic acid carbonyl warheads to covalently target a noncatalytic lysine side chain, and generated to our knowledge the first reversible covalent inhibitors for Mcl-1, a protein-protein interaction (PPI) target that has proven difficult to inhibit via traditional medicinal chemistry strategies. These covalent binders exhibited improved potency in comparison to noncovalent congeners, as demonstrated in biochemical and cell-based assays. We identified Lys234 as the residue involved in covalent modification, via point mutation. The covalent binders discovered in this study will serve as useful starting points for the development of Mcl-1 therapeutics and probes to interrogate Mcl-1-dependent biological phenomena.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Zinzalla, G. & Thurston, D.E. Targeting protein-protein interactions for therapeutic intervention: a challenge for the future. Future Med. Chem. 1, 65–93 (2009).
Azzarito, V., Long, K., Murphy, N.S. & Wilson, A.J. Inhibition of α-helix-mediated protein-protein interactions using designed molecules. Nat. Chem. 5, 161–173 (2013).
Arkin, M.R. & Wells, J.A. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 3, 301–317 (2004).
Fletcher, S. & Hamilton, A.D. Targeting protein-protein interactions by rational design: mimicry of protein surfaces. J. R. Soc. Interface 3, 215–233 (2006).
Arrowsmith, C.H. et al. The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–541 (2015).
Fischer, P.M. Protein-protein interactions in drug discovery. Drug Design Reviews Online 2, 179–207 (2005).
Way, J.C. Covalent modification as a strategy to block protein-protein interactions with small-molecule drugs. Curr. Opin. Chem. Biol. 4, 40–46 (2000).
Singh, J., Petter, R.C., Baillie, T.A. & Whitty, A. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 (2011).
Tummino, P.J. & Copeland, R.A. Residence time of receptor-ligand complexes and its effect on biological function. Biochemistry 47, 5481–5492 (2008).
Hajduk, P.J. Fragment-based drug design: how big is too big? J. Med. Chem. 49, 6972–6976 (2006).
Kuntz, I.D., Chen, K., Sharp, K.A. & Kollman, P.A. The maximal affinity of ligands. Proc. Natl. Acad. Sci. USA 96, 9997–10002 (1999).
Dutta, S. et al. Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J. Mol. Biol. 398, 747–762 (2010).
Smith, A.J.T., Zhang, X., Leach, A.G. & Houk, K.N. Beyond picomolar affinities: quantitative aspects of noncovalent and covalent binding of drugs to proteins. J. Med. Chem. 52, 225–233 (2009).
Copeland, R.A., Pompliano, D.L. & Meek, T.D. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 5, 730–739 (2006).
Walter, A.O. et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov. 3, 1404–1415 (2013).
Rudolph, J. & Stokoe, D. Selective inhibition of mutant Ras protein through covalent binding. Angew. Chem. Int. Ed. Engl. 53, 3777–3779 (2014).
Basu, D., Richters, A. & Rauh, D. Structure-based design and synthesis of covalent-reversible inhibitors to overcome drug resistance in EGFR. Bioorg. Med. Chem. 23, 2767–2780 (2015).
Finlay, M.R.V. et al. Discovery of a potent and selective EGFR inhibitor (AZD9291) of both sensitizing and T790M resistance mutations that spares the wild type form of the receptor. J. Med. Chem. 57, 8249–8267 (2014).
Barf, T. & Kaptein, A. Irreversible protein kinase inhibitors: balancing the benefits and risks. J. Med. Chem. 55, 6243–6262 (2012).
Lee, C.U. & Grossmann, T.N. Reversible covalent inhibition of a protein target. Angew. Chem. Int. Ed. Engl. 51, 8699–8700 (2012).
Serafimova, I.M. et al. Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles. Nat. Chem. Biol. 8, 471–476 (2012).
Choi, S., Connelly, S., Reixach, N., Wilson, I.A. & Kelly, J.W. Chemoselective small molecules that covalently modify one lysine in a non-enzyme protein in plasma. Nat. Chem. Biol. 6, 133–139 (2010).
Anscombe, E. et al. Identification and characterization of an irreversible inhibitor of CDK2. Chem. Biol. 22, 1159–1164 (2015).
Cal, P.M.S.D. et al. Iminoboronates: a new strategy for reversible protein modification. J. Am. Chem. Soc. 134, 10299–10305 (2012).
Bandyopadhyay, A., McCarthy, K.A., Kelly, M.A. & Gao, J. Targeting bacteria via iminoboronate chemistry of amine-presenting lipids. Nat. Commun. 6, 6561 (2015).
Verdine, G.L. & Walensky, L.D. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin. Cancer Res. 13, 7264–7270 (2007).
Cory, S. & Adams, J.M. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2, 647–656 (2002).
Czabotar, P.E., Lessene, G., Strasser, A. & Adams, J.M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 15, 49–63 (2014).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Wei, G. et al. Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell 21, 547–562 (2012).
Wenzel, S.S. et al. MCL1 is deregulated in subgroups of diffuse large B-cell lymphoma. Leukemia 27, 1381–1390 (2013).
Yancey, D. et al. BAD dephosphorylation and decreased expression of MCL-1 induce rapid apoptosis in prostate cancer cells. PLoS One 8, e74561 (2013).
Palve, V., Mallick, S., Ghaisas, G., Kannan, S. & Teni, T. Overexpression of Mcl-1L splice variant is associated with poor prognosis and chemoresistance in oral cancers. PLoS One 9, e111927 (2014).
Williams, M.M. & Cook, R.S. Bcl-2 family proteins in breast development and cancer: could Mcl-1 targeting overcome therapeutic resistance? Oncotarget 6, 3519–3530 (2015).
Ashkenazi, A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat. Rev. Drug Discov. 7, 1001–1012 (2008).
Yamanaka, K. et al. A novel antisense oligonucleotide inhibiting several antiapoptotic Bcl-2 family members induces apoptosis and enhances chemosensitivity in androgen-independent human prostate cancer PC3 cells. Mol. Cancer Ther. 4, 1689–1698 (2005).
Cohen, N.A. et al. A competitive stapled peptide screen identifies a selective small molecule that overcomes MCL-1-dependent leukemia cell survival. Chem. Biol. 19, 1175–1186 (2012).
Muppidi, A. et al. Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors. J. Am. Chem. Soc. 134, 14734–14737 (2012).
Tanaka, Y. et al. Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins. J. Med. Chem. 56, 9635–9645 (2013).
Friberg, A. et al. Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design. J. Med. Chem. 56, 15–30 (2013).
AMG 176 First in Human Trial in Subjects With Relapsed or Refractory Multiple Myeloma. https://clinicaltrials.gov/ct2/show/NCT02675452.
Bruncko, M., Song, X., Ding, H., Tao, Z.F. & Kunzer, A.R. 7-nonsubstituted indole mcl-1 inhibitors. WO Patent 2008130970 A1 filed 28 April 2008, and issued 2 March 2016.
Bruncko, M. et al. Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity. J. Med. Chem. 58, 2180–2194 (2015).
Belmonte, M.A. et al. Evaluation of Mcl-1 inhibitors in preclinical models of multiple myeloma. Blood 124, 3428 (2014).
Touzeau, C. et al. BH3 profiling identifies heterogeneous dependency on Bcl-2 family members in multiple myeloma and predicts sensitivity to BH3 mimetics. Leukemia 30, 761–764 (2016).
Booher, R.N. et al. MCL1 and BCL-xL levels in solid tumors are predictive of dinaciclib-induced apoptosis. PLoS One 9, e108371 (2014).
Cal, P.M.S.D., Frade, R.F.M., Cordeiro, C. & Gois, P.M.P. Reversible lysine modification on proteins by using functionalized boronic acids. Chemistry 21, 8182–8187 (2015).
Mah, R., Thomas, J.R. & Shafer, C.M. Drug discovery considerations in the development of covalent inhibitors. Bioorg. Med. Chem. Lett. 24, 33–39 (2014).
Belmar, J. & Fesik, S.W. Small molecule Mcl-1 inhibitors for the treatment of cancer. Pharmacol. Ther. 145, 76–84 (2015).
Mukherjee, H. et al. Interactions of the natural product (+)-avrainvillamide with nucleophosmin and exportin-1 mediate the cellular localization of nucleophosmin and its AML-associated mutants. ACS Chem. Biol. 10, 855–863 (2015).
Kenny, P.W. & Sadowski, J. Structure modification in chemical databases. in Chemoinformatics in Drug Discovery (ed., T.I. Oprea) Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, FRG) 271–285 (2005).
Gasteiger, J., Rudolph, C. & Sadowski, J. Automatic generation of 3D-atomic coordinates for organic molecules. Tetrahedron Computer Methodology 3, 537–547 (1990).
Acknowledgements
We thank P. Ross for protein mass spectrometry assistance, E. Code and K. Jacques for advice and help for the preparation of Mcl-1 K234A mutant, A. Shapiro and J. Breen for discussion of the kinetic data obtained from time-dependent TR-FRET binding experiments, R. Chen for helpful discussions, and M. Vasbinder for proofreading the manuscript and helpful discussions. We thank the AstraZeneca PostDoc Program for funding this project.
Author information
Authors and Affiliations
Contributions
G.A., N.P.G., B.A., C.C., A.W.H., M.L.L. and Q.S. designed the covalent inhibitors; G.A. and N.P.G. synthesized all compounds; G.A., M.A.B., P.B.R. and N.S. performed biological experiments. G.A., N.P.G., C.C., M.A.B., P.B.R., M.L.L., A.W.H., B.A., N.S., S.T. and Q.S. interpreted and discussed the results. G.A. and N.P.G. wrote the manuscript. All authors contributed to editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors are current or former employees of AstraZeneca Pharmaceuticals.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–11 and Supplementary Tables 1–4. (PDF 1331 kb)
Supplementary Note
Synthetic Procedures (PDF 849 kb)
Rights and permissions
About this article
Cite this article
Akçay, G., Belmonte, M., Aquila, B. et al. Inhibition of Mcl-1 through covalent modification of a noncatalytic lysine side chain. Nat Chem Biol 12, 931–936 (2016). https://doi.org/10.1038/nchembio.2174
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2174
This article is cited by
-
A chemical proteomics approach for global mapping of functional lysines on cell surface of living cell
Nature Communications (2024)
-
A novel Mcl-1 inhibitor synergizes with venetoclax to induce apoptosis in cancer cells
Molecular Medicine (2023)
-
An update on the discovery and development of reversible covalent inhibitors
Medicinal Chemistry Research (2023)
-
Promising reversible protein inhibitors kept on target
Nature (2022)
-
Discovery and molecular basis of subtype-selective cyclophilin inhibitors
Nature Chemical Biology (2022)