Abstract
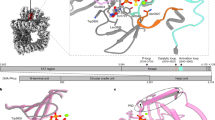
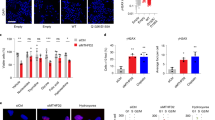
The structure-specific nuclease human flap endonuclease-1 (hFEN1) plays a key role in DNA replication and repair and may be of interest as an oncology target. We present the crystal structure of inhibitor-bound hFEN1, which shows a cyclic N-hydroxyurea bound in the active site coordinated to two magnesium ions. Three such compounds had similar IC50 values but differed subtly in mode of action. One had comparable affinity for protein and protein–substrate complex and prevented reaction by binding to active site catalytic metal ions, blocking the necessary unpairing of substrate DNA. Other compounds were more competitive with substrate. Cellular thermal shift data showed that both inhibitor types engaged with hFEN1 in cells, and activation of the DNA damage response was evident upon treatment with inhibitors. However, cellular EC50 values were significantly higher than in vitro inhibition constants, and the implications of this for exploitation of hFEN1 as a drug target are discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Grasby, J.A., Finger, L.D., Tsutakawa, S.E., Atack, J.M. & Tainer, J.A. Unpairing and gating: sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem. Sci. 37, 74–84 (2012).
Tsutakawa, S.E. et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145, 198–211 (2011).
Kim, C.Y., Shen, B., Park, M.S. & Olah, G.A. Structural changes measured by X-ray scattering from human flap endonuclease-1 complexed with Mg2+ and flap DNA substrate. J. Biol. Chem. 274, 1233–1239 (1999).
Zheng, L. et al. Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res. 39, 781–794 (2011).
Waga, S., Bauer, G. & Stillman, B. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 269, 10923–10934 (1994).
Bambara, R.A., Murante, R.S. & Henricksen, L.A. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272, 4647–4650 (1997).
Kim, K., Biade, S. & Matsumoto, Y. Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem. 273, 8842–8848 (1998).
Parikh, S.S., Mol, C.D., Hosfield, D.J. & Tainer, J.A. Envisioning the molecular choreography of DNA base excision repair. Curr. Opin. Struct. Biol. 9, 37–47 (1999).
Beard, W.A. & Wilson, S.H. Structure and mechanism of DNA polymerase Beta. Chem. Rev. 106, 361–382 (2006).
Mohan, V. & Srinivasan, M. in New Research Directions in DNA Repair (ed. Clark Chen) (InTech, 2013).
Singh, P. et al. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol. Cancer Res. 6, 1710–1717 (2008).
Lam, J.S. et al. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 98, 445–451 (2006).
Nikolova, T., Christmann, M. & Kaina, B. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res. 29, 2453–2459 (2009).
Panda, H. et al. Amino acid Asp181 of 5′-flap endonuclease 1 is a useful target for chemotherapeutic development. Biochemistry 48, 9952–9958 (2009).
Yoshimoto, K. et al. Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front. Oncol. 2, 186 (2012).
van Pel, D.M. et al. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 9, e1003254 (2013).
Illuzzi, J.L. & Wilson, D.M. III. Base excision repair: contribution to tumorigenesis and target in anticancer treatment paradigms. Curr. Med. Chem. 19, 3922–3936 (2012).
McManus, K.J., Barrett, I.J., Nouhi, Y. & Hieter, P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc. Natl. Acad. Sci. USA 106, 3276–3281 (2009).
Durant, S.T. Telomerase-independent paths to immortality in predictable cancer subtypes. J. Cancer 3, 67–82 (2012).
Hwang, J.-C. et al. The overexpression of FEN1 and RAD54B may act as independent prognostic factors of lung adenocarcinoma. PLoS One 10, e0139435 (2015).
Shibata, Y. & Nakamura, T. Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J. Biol. Chem. 277, 746–754 (2002).
McWhirter, C. et al. Development of a high-throughput fluorescence polarization DNA cleavage assay for the identification of FEN1 inhibitors. J. Biomol. Screen. 18, 567–575 (2013).
Dorjsuren, D., Kim, D., Maloney, D.J., Wilson, D.M. III & Simeonov, A. Complementary non-radioactive assays for investigation of human flap endonuclease 1 activity. Nucleic Acids Res. 39, e11 (2011).
Tumey, L.N. et al. The identification and optimization of a N-hydroxy urea series of flap endonuclease 1 inhibitors. Bioorg. Med. Chem. Lett. 15, 277–281 (2005).
Finger, L.D. et al. The wonders of flap endonucleases: structure, function, mechanism and regulation. Subcell. Biochem. 62, 301–326 (2012).
Guo, Z. et al. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol. Cell 47, 444–456 (2012).
Sakurai, S. et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 24, 683–693 (2005).
Finger, L.D. et al. Observation of unpaired substrate DNA in the flap endonuclease-1 active site. Nucleic Acids Res. 41, 9839–9847 (2013).
Patel, N. et al. Flap endonucleases pass 5′-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5′-ends. Nucleic Acids Res. 40, 4507–4519 (2012).
Syson, K. et al. Three metal ions participate in the reaction catalyzed by T5 flap endonuclease. J. Biol. Chem. 283, 28741–28746 (2008).
Tomlinson, C.G. et al. Neutralizing mutations of carboxylates that bind metal 2 in T5 flap endonuclease result in an enzyme that still requires two metal ions. J. Biol. Chem. 286, 30878–30887 (2011).
Finger, L.D. et al. The 3′-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis. J. Biol. Chem. 284, 22184–22194 (2009).
Patel, N. et al. Proline scanning mutagenesis reveals a role for the flap endonuclease-1 helical cap in substrate unpairing. J. Biol. Chem. 288, 34239–34248 (2013).
Craggs, T.D., Hutton, R.D., Brenlla, A., White, M.F. & Penedo, J.C. Single-molecule characterization of Fen1 and Fen1/PCNA complexes acting on flap substrates. Nucleic Acids Res. 42, 1857–1872 (2014).
Orans, J. et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145, 212–223 (2011).
Niesen, F.H., Berglund, H. & Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 (2007).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Moldovan, G.L. & D'Andrea, A.D. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 43, 223–249 (2009).
Yeo, J.E., Lee, E.H., Hendrickson, E.A. & Sobeck, A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum. Mol. Genet. 23, 3695–3705 (2014).
Schlacher, K., Wu, H. & Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012).
Di Santo, R. Inhibiting the HIV integration process: past, present, and the future. J. Med. Chem. 57, 539–566 (2014).
Dahl, G. & Akerud, T. Pharmacokinetics and the drug-target residence time concept. Drug Discov. Today 18, 697–707 (2013).
Fehrmann, R.S.N. et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 47, 115–125 (2015).
Bolderson, E. et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 38, 1821–1831 (2010).
Desai, A., Qing, Y. & Gerson, S.L. Exonuclease 1 is a critical mediator of survival during DNA double strand break repair in nonquiescent hematopoietic stem and progenitor cells. Stem Cells 32, 582–593 (2014).
Tomimatsu, N. et al. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat. Commun. 5, 3561 (2014).
Tomimatsu, N. et al. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst.) 11, 441–448 (2012).
Eschenfeldt, W.H. et al. Cleavable C-terminal His-tag vectors for structure determination. J. Struct. Funct. Genomics 11, 31–39 (2010).
Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Fien, K. & Stillman, B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell. Biol. 12, 155–163 (1992).
Chang, J.H., Xiang, S., Xiang, K., Manley, J.L. & Tong, L. Structural and biochemical studies of the 5′→3′ exoribonuclease Xrn1. Nat. Struct. Mol. Biol. 18, 270–276 (2011).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Keller, S. et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 (2012).
Houtman, J.C.D. et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 (2007).
Izumi, T. & Mitra, S. Deletion analysis of human AP-endonuclease: minimum sequence required for the endonuclease activity. Carcinogenesis 19, 525–527 (1998).
Clegg, R.M. et al. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry 31, 4846–4856 (1992).
Acknowledgements
This work was supported by U.K. Biotechnology and Biological Sciences Research Council grants BB/K009079/1 and BB/M00404X/1 (both to J.A.G.) and AstraZeneca. J.C.E. thanks the U.K. Engineering and Physical Sciences Research Council and AstraZeneca for a studentship. The authors thank C. Phillips for assistance with submissions of the crystallographic data and T. McGuire for synthetic support.
Author information
Authors and Affiliations
Contributions
C.D.J. designed and synthesized inhibitors. J.C.E., M.J.T., L.D.F. and S.J.S. carried out kinetic and biophysical experiments. J.C.E., M.J.T., L.D.F., C.M., J.W.M.N., W.M.A. and J.A.G. designed experiments and analyzed this data. J.D. and J.W.M.N. obtained and analyzed structures. C.L.B.S. and D.M.M. performed the CETSA assays. T.A.W. carried out other cellular experiments, and T.A.W. and S.T.D. analyzed data. All authors contributed to the preparation of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
W.M.A., C.M., J.D., J.W.M.N. and S.T.D. are employees of AstraZeneca. C.D.J. and T.A.W. were employees of AstraZeneca at the time of writing. C.L.B.S. and D.M.M. are employees of Pelago Bioscience AB.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–6 and Supplementary Figures 1–20. (PDF 8959 kb)
Supplementary Note
Synthetic Procedures (PDF 511 kb)
Rights and permissions
About this article
Cite this article
Exell, J., Thompson, M., Finger, L. et al. Cellularly active N-hydroxyurea FEN1 inhibitors block substrate entry to the active site. Nat Chem Biol 12, 815–821 (2016). https://doi.org/10.1038/nchembio.2148
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2148
This article is cited by
-
FEN1 inhibitor synergizes with low-dose camptothecin to induce increased cell killing via the mitochondria mediated apoptotic pathway
Gene Therapy (2022)
-
Metal–ligand interactions in drug design
Nature Reviews Chemistry (2018)
-
Phosphate steering by Flap Endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability
Nature Communications (2017)