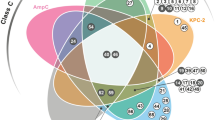
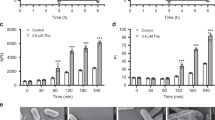
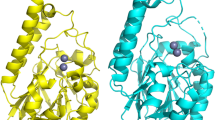
Abstract
Production of β-lactamases of one of four molecular classes (A, B, C and D) is the major mechanism of bacterial resistance to β-lactams, the largest class of antibiotics, which have saved countless lives since their inception 70 years ago. Although several hundred efficient class D enzymes have been identified in Gram-negative pathogens over the last four decades, none have been reported in Gram-positive bacteria. Here we demonstrate that efficient class D β-lactamases capable of hydrolyzing a wide array of β-lactam substrates are widely disseminated in various species of environmental Gram-positive organisms. Class D enzymes of Gram-positive bacteria have a distinct structural architecture and employ a unique substrate-binding mode that is quite different from that of all currently known class A, C and D β-lactamases. These enzymes thus constitute a previously unknown reservoir of novel antibiotic-resistance enzymes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Referenced accessions
GenBank/EMBL/DDBJ
NCBI Reference Sequence
Protein Data Bank
References
Poole, K. Resistance to β-lactam antibiotics. Cell. Mol. Life Sci. 61, 2200–2223 (2004).
Waxman, D.J. & Strominger, J.L. Penicillin-binding proteins and the mechanism of action of β-lactam antibiotics. Annu. Rev. Biochem. 52, 825–869 (1983).
Macheboeuf, P., Contreras-Martel, C., Job, V., Dideberg, O. & Dessen, A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30, 673–691 (2006).
Cho, H., Uehara, T. & Bernhardt, T.G. β-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311 (2014).
Bush, K. Proliferation and significance of clinically relevant β-lactamases. Ann. NY Acad. Sci. 1277, 84–90 (2013).
Ambler, R.P. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci 289, 321–331 (1980).
Jaurin, B. & Grundstrom, T. AmpC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamases of the penicillinase type. Proc. Natl. Acad. Sci. USA 78, 4897–4901 (1981).
Ouellette, M., Bissonnette, L. & Roy, P.H. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 β-lactamase gene. Proc. Natl. Acad. Sci. USA 84, 7378–7382 (1987).
Kernodle, D.S. in Gram-positive Pathogens (eds. Fischetti, V.A., Novick, R.P., Ferretti, J.J., Portnoy, D.A. & Rood, J.I.) 609–620 (ASM Press, Washington, D.C., 2000).
Hedges, R.W., Datta, N., Kontomichalou, P. & Smith, J.T. Molecular specificities of R factor-determined β-lactamases: Correlation with plasmid compatibility. J. Bacteriol. 117, 56–62 (1974).
Sykes, R.B. & Matthew, M. The β-lactamases of gram-negative bacteria and their role in resistance to β-lactam antibiotics. J. Antimicrob. Chemother. 2, 115–157 (1976).
Leonard, D.A., Bonomo, R.A. & Powers, R.A. Class D β-lactamases: a reappraisal after five decades. Acc. Chem. Res. 46, 2407–2415 (2013).
Walther-Rasmussen, J. & Hoiby, N. OXA-type carbapenemases. J. Antimicrob. Chemother. 57, 373–383 (2006).
Poirel, L., Naas, T. & Nordmann, P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 54, 24–38 (2010).
Antunes, N.T. et al. Class D β-lactamases: are they all carbapenemases? Antimicrob. Agents Chemother. 58, 2119–2125 (2014).
Docquier, J.D. et al. Crystal structure of the OXA-48 β-lactamase reveals mechanistic diversity among class D carbapenemases. Chem. Biol. 16, 540–547 (2009).
Verma, V. et al. Hydrolytic mechanism of OXA-58 enzyme, a carbapenem-hydrolyzing class D β-lactamase from Acinetobacter baumannii. J. Biol. Chem. 286, 37292–37303 (2011).
Colombo, M.-L. et al. The ybxI gene of Bacillus subtilis 168 encodes a class D β-lactamase of low activity. Antimicrob. Agents Chemother. 48, 484–490 (2004).
Docquier, J.D. et al. Crystal structure of the narrow-spectrum OXA-46 class D β-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates. Antimicrob. Agents Chemother. 54, 2167–2174 (2010).
Golemi, D. et al. The first structural and mechanistic insights for class D β-lactamases: evidence for a novel catalytic process for turnover of β-lactam antibiotics. J. Am. Chem. Soc. 122, 6132–6133 (2000).
Kaitany, K.C. et al. Structures of the class D carbapenemases OXA-23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins and aztreonam. Antimicrob. Agents Chemother. 57, 4848–4855 (2013).
Paetzel, M. et al. Crystal structure of the class D β-lactamase OXA-10. Nat. Struct. Biol. 7, 918–925 (2000).
Pernot, L. et al. Crystal structures of the class D β-lactamase OXA-13 in the native form and in complex with meropenem. J. Mol. Biol. 310, 859–874 (2001).
Santillana, E., Beceiro, A., Bou, G. & Romero, A. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. USA 104, 5354–5359 (2007).
Smith, C.A. et al. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii. Chem. Biol. 20, 1107–1115 (2013).
Smith, C.A., Antunes, N.T., Toth, M. & Vakulenko, S.B. Crystal structure of carbapenemase OXA-58 from Acinetobacter baumannii. Antimicrob. Agents Chemother. 58, 2135–2143 (2014).
Sun, T., Nukaga, M., Mayama, K., Braswell, E.H. & Knox, J.R. Comparison of β-lactamases of classes A and D: 1.5-Å crystallographic structure of the class D OXA-1 oxacillinase. Protein Sci. 12, 82–91 (2003).
Golemi, D., Maveyraud, L., Vakulenko, S., Samama, J.P. & Mobashery, S. Critical involvement of a carbamylated lysine in catalytic function of class D β-lactamases. Proc. Natl. Acad. Sci. USA 98, 14280–14285 (2001).
Schneider, K.D. et al. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumanii in complex with doripenem. J. Mol. Biol. 406, 583–594 (2011).
Beadle, B.M. & Shoichet, B.K. Structural basis for imipenem inhibition of class C β-lactamases. Antimicrob. Agents Chemother. 46, 3978–3980 (2002).
Ambler, R.P. et al. A standard numbering scheme for the class-A β-lactamases. Biochem. J. 276, 269–270 (1991).
Marciano, D.C., Brown, N.G. & Palzkill, T. Analysis of the plasticity of location of the Arg244 positive charge within the active site of the TEM-1 β-lactamase. Protein Sci. 18, 2080–2089 (2009).
Giannouli, M. et al. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J. Clin. Microbiol. 48, 1223–1230 (2010).
Curley, K. & Pratt, R.F. The oxyanion hole in serine β-lactamase catalysis: interactions of thiono substrates with the active site. Bioorg. Chem. 28, 338–356 (2000).
Maveyraud, L. et al. Crystal structure of 6α-(hydroxymethyl)penicillanate complexed to the TEM-1 β-lactamase from Escherichia coli: evidence on the mechanism of action of a novel inhibitor designed by a computer-aided process. J. Am. Chem. Soc. 118, 7435–7440 (1996).
Mourey, L. et al. Inhibition of the NMC-A β-lactamase by a penicillanic acid derivative, and the structural bases for the increase in substrate profile of this antibiotic resistance enzyme. J. Am. Chem. Soc. 120, 9382–9383 (1998).
Patera, A., Blaszczak, L.C. & Shoichet, B.K. Crystal structures of substrate and inhibitor complexes with AmpC β-lactamase: possible implications for substrate-assisted catalysis. J. Am. Chem. Soc. 122, 10504–10512 (2000).
Powers, R.A., Caselli, E., Focia, P.J., Prati, F. & Shoichet, B.K. Structures of ceftazidime and its transition-state analogue in complex with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40, 9207–9214 (2001).
Stewart, N.K., Smith, C.A., Frase, H., Black, D.J. & Vakulenko, S.B. Kinetic and structural requirements for carbapenemase activity in GES-type β-lactamases. Biochemistry 54, 588–597 (2015).
Schneider, K.D., Karpen, M.E., Bonomo, R.A., Leonard, D.A. & Powers, R.A. The 1.4 Å crystal structure of the class D β-lactamase OXA-1 complexed with doripenem. Biochemistry 48, 11840–11847 (2009).
Thomson, J.M., Distler, A.M., Prati, F. & Bonomo, R.A. Probing active site chemistry in SHV β-lactamase variants at Ambler position 244. Understanding unique properties of inhibitor resistance. J. Biol. Chem. 281, 26734–26744 (2006).
Wattam, A.R. et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42, D581–D591 (2014).
Toth, M., Smith, C., Frase, H., Mobashery, S. & Vakulenko, S. An antibiotic-resistance enzyme from a deep-sea bacterium. J. Am. Chem. Soc. 132, 816–823 (2010).
Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard (Clinical and Laboratory Standards Institute, Wayne, PA, 2009).
Matthews, B.W. Solvent contents of protein crystals. J. Mol. Biol. 33, 491–497 (1968).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Evans, P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 (2011).
Evans, P.R. & Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Murshudov, G.N., Vagin, A.A., Lebedev, A., Wilson, K.S. & Dodson, E.J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 (1999).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
The Stanford Synchrotron Radiation Lightsource (SSRL) is a national user facility operated by Stanford University and supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the US National Institutes of Health (NIH), National Institute of General Medical Sciences (NIGMS) (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Author information
Authors and Affiliations
Contributions
S.B.V. and N.T.A. performed the genomic sequence screening and sequence alignments; M.T., N.T.A., N.K.S., M.B. and H.F. designed and performed the microbiological and kinetic experiments and analyzed the resulting data; M.T., N.T.A. and C.A.S. designed and performed the crystallography experiments; C.A.S. collected and analyzed the crystallographic data; S.B.V. and C.A.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–4 and Supplementary Tables 1–3. (PDF 1350 kb)
Rights and permissions
About this article
Cite this article
Toth, M., Antunes, N., Stewart, N. et al. Class D β-lactamases do exist in Gram-positive bacteria. Nat Chem Biol 12, 9–14 (2016). https://doi.org/10.1038/nchembio.1950
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1950