Abstract
Transposons have had a pivotal role in genome evolution1 and are believed to be the evolutionary progenitors of the RAG1–RAG2 recombinase2, an essential component of the adaptive immune system in jawed vertebrates3. Here we report one crystal structure and five cryo-electron microscopy structures of Transib4,5, a RAG1-like transposase from Helicoverpa zea, that capture the entire transposition process from the apo enzyme to the terminal strand transfer complex with transposon ends covalently joined to target DNA, at resolutions of 3.0–4.6 Å. These structures reveal a butterfly-shaped complex that undergoes two cycles of marked conformational changes in which the ‘wings’ of the transposase unfurl to bind substrate DNA, close to execute cleavage, open to release the flanking DNA and close again to capture and attack target DNA. Transib possesses unique structural elements that compensate for the absence of a RAG2 partner, including a loop that interacts with the transposition target site and an accordion-like C-terminal tail that elongates and contracts to help to control the opening and closing of the enzyme and assembly of the active site. Our findings reveal the detailed reaction pathway of a eukaryotic cut-and-paste transposase and illuminate some of the earliest steps in the evolution of the RAG recombinase.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Atomic coordinates of six HzTransib or HzTransib–TIR DNA complex structures have been deposited in PDB under accession number 6PQN (HzTransib apo), 6PQR (HzTransib intact TIR PRC), 6PQU (HzTransib nicked TIR PRC), 6PQX (HzTransib TIR HFC), 6PQY (HzTransib TIR TEC) and 6PR5 (HzTransib TIR STC). Five cryo-EM density maps of HzTransib complexed with different TIR DNA have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-20452, EMD-20453, EMD-20455, EMD-20456 and EMD-20457, respectively.
References
Feschotte, C. & Pritham, E. J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368 (2007).
Carmona, L. M. & Schatz, D. G. New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination. FEBS J. 284, 1590–1605 (2017).
Gellert, M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71, 101–132 (2002).
Chen, S. & Li, X. Molecular characterization of the first intact Transib transposon from Helicoverpa zea. Gene 408, 51–63 (2008).
Hencken, C. G., Li, X. & Craig, N. L. Functional characterization of an active Rag-like transposase. Nat. Struct. Mol. Biol. 19, 834–836 (2012).
Craig, N. L. in Mobile DNA III (eds Craig, N. L. et al.) 3–39 (ASM Press, 2015).
Montaño, S. P. & Rice, P. A. Moving DNA around: DNA transposition and retroviral integration. Curr. Opin. Struct. Biol. 21, 370–378 (2011).
Kim, M. S., Lapkouski, M., Yang, W. & Gellert, M. Crystal structure of the V(D)J recombinase RAG1–RAG2. Nature 518, 507–511 (2015).
Schatz, D. G. & Swanson, P. C. V. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45, 167–202 (2011).
Kapitonov, V. V. & Jurka, J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 3, e181 (2005).
Zhou, L. et al. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature 432, 995–1001 (2004).
Hickman, A. B. et al. Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell 158, 353–367 (2014).
Agrawal, A., Eastman, Q. M. & Schatz, D. G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–751 (1998).
Hiom, K., Melek, M. & Gellert, M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94, 463–470 (1998).
Ru, H. et al. Molecular mechanism of V(D)J recombination from synaptic RAG1–RAG2 complex structures. Cell 163, 1138–1152 (2015).
Kim, M. S. et al. Cracking the DNA code for V(D)J recombination. Mol. Cell 70, 358–370 (2018).
Ru, H. et al. DNA melting initiates the RAG catalytic pathway. Nat. Struct. Mol. Biol. 25, 732–742 (2018).
Montaño, S. P., Pigli, Y. Z. & Rice, P. A. The μ transpososome structure sheds light on DDE recombinase evolution. Nature 491, 413–417 (2012).
Maertens, G. N., Hare, S. & Cherepanov, P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468, 326–329 (2010).
Yin, Z. et al. Crystal structure of the Rous sarcoma virus intasome. Nature 530, 362–366 (2016).
Ballandras-Colas, A. et al. A supramolecular assembly mediates lentiviral DNA integration. Science 355, 93–95 (2017).
Passos, D. O. et al. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 355, 89–92 (2017).
Richardson, J. M., Colloms, S. D., Finnegan, D. J. & Walkinshaw, M. D. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell 138, 1096–1108 (2009).
Morris, E. R., Grey, H., McKenzie, G., Jones, A. C. & Richardson, J. M. A bend, flip and trap mechanism for transposon integration. eLife 5, e15537 (2016).
Dawson, A. & Finnegan, D. J. Excision of the Drosophila mariner transposon Mos1. Comparison with bacterial transposition and V(D)J recombination. Mol. Cell 11, 225–235 (2003).
Carmona, L. M., Fugmann, S. D. & Schatz, D. G. Collaboration of RAG2 with RAG1-like proteins during the evolution of V(D)J recombination. Genes Dev. 30, 909–917 (2016).
Davies, D. R., Goryshin, I. Y., Reznikoff, W. S. & Rayment, I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289, 77–85 (2000).
Lankaš, F., Sponer, J., Langowski, J. & Cheatham, T. E. III. DNA basepair step deformability inferred from molecular dynamics simulations. Biophys. J. 85, 2872–2883 (2003).
Yuan, Y. W. & Wessler, S. R. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl Acad. Sci. USA 108, 7884–7889 (2011).
Hickman, A. B. et al. Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase. Nucleic Acids Res. 46, 10286–10301 (2018).
Yang, W., Lee, J. Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006).
Huang, S. et al. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell 166, 102–114 (2016).
Zhang, Y. et al. Transposon molecular domestication and the evolution of the RAG recombinase. Nature 569, 79–84 (2019).
Hare, S., Gupta, S. S., Valkov, E., Engelman, A. & Cherepanov, P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236 (2010).
Tsai, C. L., Chatterji, M. & Schatz, D. G. DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res. 31, 6180–6190 (2003).
Lee, G. S., Neiditch, M. B., Sinden, R. R. & Roth, D. B. Targeted transposition by the V(D)J recombinase. Mol. Cell. Biol. 22, 2068–2077 (2002).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D 65, 582–601 (2009).
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D 64, 61–69 (2008).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Sievers, F. & Higgins, D. G. Clustal omega. Curr. Protoc. Bioinformatics 48, 3.13.1–3.13.16 (2014).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Wu, X. & Bartel, D. P. kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res. 45, W534–W538 (2017).
Acknowledgements
We thank W. Eliason for assistance with size-exclusion chromatography–multiple angle light scattering; K. Zhou for assistance in freezing the cryo-EM grids of HzTransib-intact TIR complex; S. Wu for help with cryo-EM data collection at Yale West Campus; the staff of the Advanced Photon Source beamlines 24-ID-C and 24-ID-E for technical assistance with X-ray crystallography data collection; N. Craig for critical reading and many helpful comments on the manuscript. We are grateful for the advice, mentoring and support from T. Steitz during the early phases of this work. This work was supported by NIH grant R01 AI137079 (D.G.S.), Yale University School of Medicine James Hudson Brown-Alexander Brown Coxe Postdoctoral Fellowship (C.L.) and NVIDIA GPU Grant Program (C.L. and Y.Y.).
Author information
Authors and Affiliations
Contributions
C.L. and D.G.S. conceived the project and designed the experiments. C.L. performed cloning, protein expression, purification, complex reconstitution, sample screening using negative-stain electron microscopy, cryo-EM grids preparation and functional assays. C.L. and Y.Y. carried out protein crystallization, crystal structure determination, cryo-EM data collection and processing, atomic model building and refinement. C.L. and D.G.S. analysed data and wrote the manuscript with input from Y.Y.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Orsolya Barabas, Thomas Boehm, Ronald Chalmers and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
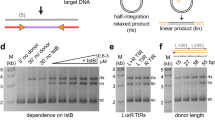
Extended Data Fig. 1 Biochemical characterization of H. zea Transib transposase and single-particle cryo-EM analysis of Transib in complex with intact TIR substrates.
a, Size-exclusion chromatography–multiple angle light scattering analysis of purified Transib protein, indicating that it forms a dimer in solution. Size-exclusion chromatography was repeated three times and similar profiles were obtained. The multiple angle light scattering experiment was not repeated. b, Numbering and sequence of endogenous left end (5′TIR) and right end (3′TIR) of the Transib transposon with nucleotide differences in black boxes. The first 16 bp of the TIR sequence are the same as the 16-bp transposon end of the TIR substrates used in structure determination. c, Schematic of the TIR substrate DNA used in the in vitro DNA-cleavage assay. 5′TIR and 3′TIR are shown as yellow and purple triangles, respectively. d, Cleavage of DNA substrates bearing one or two TIRs by MBP-tagged wild-type or mutant Transib transposases, each with the N-terminal 16 amino acids removed. The experiment was repeated three times and similar results were obtained. For gel source data, see Supplementary Fig. 1. e, Cleavage of DNA substrates bearing either full-length (lanes 1 and 2) or truncated (lanes 3–8) 5′TIR or 3′TIR, with site of truncation indicated in the substrate name. The experiment was repeated three times and similar results were obtained. Open and closed arrowheads indicate single 5′TIR and single 3′TIR cleavage products, respectively. Red asterisk marks the double cleavage band. The DNA cleavage products were resolved in 5% TBE polyacrylamide gels and stained with SYBR Gold. f, Flow chart of cryo-EM structure determination of Transib in complex with intact TIR substrates. After the first round of 3D classification, 3D auto-refinement using all of the particles in the best class generated a 3.3 Å map. Further 3D classifications focusing on either two ZnB domains plus flanking DNA regions or on one ZnB domain with symmetry expansion were used to obtain the final 3.4 Å map or a 3.5 Å map with clear ZnB domain density. All three maps were used as cross-references for model building. The final map and accompanying local resolution illustrations are enclosed in the dashed black box.
Extended Data Fig. 2 Single-particle cryo-EM analysis of Transib in complex with nicked TIR substrates.
a, Flow chart of cryo-EM image processing for Transib PRC with nicked TIR substrates. After the first round of 3D classification with two MBP regions masked, 3D auto-refinement using all of the particles in the best class generated a 3.2 Å map. Further 3D classifications focusing on either two ZnB domains plus flanking DNA regions or on one ZnB domain with symmetry expansion were used to obtain the final 3.3 Å map or a 3.4 Å map with clear ZnB domain density. All three maps were used as cross-references for model building. b, Flow chart of cryo-EM image processing for Transib HFC with nicked TIR substrates. After initial 3D auto-refinement, particles in the over-represented 2D classes were manually adjusted to alleviate the preferred particle orientation problem. Subsequent 3D classification and auto-refinement yielded a 4.6 Å map with even angular distribution. The final maps and accompanying local resolution illustrations are enclosed in the dashed black box.
Extended Data Fig. 3 Single-particle cryo-EM analysis of Transib in complex with TIR substrates in reaction conditions that support catalysis.
Flow chart for Transib TEC and STC map reconstructions from Transib–intact TIR DNA complex prepared at 30 °C in the presence of Mg2+. Different subsets of particle images were selected from different classification schemes to produce three refined cryo-EM maps: final STC map at 3.3 Å, final TEC map at 4.2 Å and a 4.5 Å map encompassing one Transib and TIR protomer in TEC with clear ZnB domain density. The two TEC maps were used as cross-references for model building. The final STC and TEC maps and accompanying local resolution illustrations are enclosed in the dashed black box.
Extended Data Fig. 4 Validation of cryo-EM structural models.
a, Half-map FSC and model-map FSC curves of five cryo-EM maps from this study were generated from MolProbity. Gold-standard FSC curves between the two half maps with indicated resolution at FSC = 0.143 are in blue. FSC curves between the atomic model and the final map with indicated resolution at FSC = 0.5 are in orange. b, Cryo-EM densities superimposed on the atomic model for representative regions of Transib and TIR complexes.
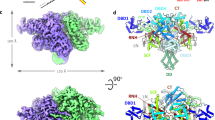
Extended Data Fig. 5 Structural comparison of Transib with RAG1.
a, Superimposition of individual domains from Transib and RAG1 structures. Because the ZnC2 portion of the ZnB domain is missing from the Transib apo structure, the ZnB domain from Transib STC was used for structural superimposition. Three structural motifs in RAG1 that are responsible for RAG2 interactions are highlighted in red boxes. b, The front and top views of Transib and RAG1 dimer superimposed by their DDBD domains. c, Front and top view of the apo RAG1-RAG2 heterotetramer structure (PDB 4WWX)8.
Extended Data Fig. 6 TIR recognition in Transib PRC, HFC and TEC.
a, Superimposition of Transib dimer in PRC (dark colours) and apo (pale colours) structures by their DDBD illustrates the large conformational changes of ZnB domains (green in one subunit). b–e, TIR recognition in Transib PRC. b, Interactions between Transib CTT and the heptamer. Hydrogen bonds are shown as grey dotted lines. Labels for nucleotide residues are italic. c, Interactions between Transib and last three base pairs of heptamer. d, Interactions between Transib and transposon end DNA downstream of heptamer. e, Active site of Transib PRC structure. Distances between Mg2+ ion and scissile phosphate or E435 are indicated. f, The front and top views of two Transib PRC structures (incubated with either intact or nicked TIRs at 4 °C) superimposed by their DDBD domains. The Transib nicked PRC complex is referred to as a PRC because of its strong structural resemblance to the intact DNA PRC. Depending on reaction conditions (temperature and divalent cation; see Methods), the nicked TIR substrate can be incorporated into either a nicked PRC or the HFC. g, Superimposition of Transib dimer in HFC and PRC structures by their DDBD shows the inward movements of ZnB domains and dimer closure. h–k, TIR recognition in Transib HFC. h, Interactions between Transib and the first three base pairs of heptamer. i, The first nucleotide of the heptamer (C1) is flipped out and buried in a pocket. j, Interactions between Transib α9–α10 loop and TIR at heptamer-flanking DNA junction. k, Active site of Transib HFC structure. l, Interactions between Transib and TIR flanking DNA in PRC. m, Interactions between Transib ZnB domain and TIR flanking DNA in HFC. n, Superimposition of Transib dimer in TEC and HFC structures by their DDBD shows the outward movements of ZnB domains. o, Comparison of transposon end DNA in TEC to that in HFC or in PRC. Mg2+ and Ca2+ ions are green and slate grey, respectively; other structure elements are coloured as in Fig. 2b. Scissile phosphate in each structure is highlighted in yellow.
Extended Data Fig. 7 Validation and analysis of Transib STC structure.
a, Superimposition of 5-bp TSD region with the cryo-EM map contoured at 5.5σ. b, Front and top views of Transib STC structure superimposed on the Transib HFC structure. c, Comparison of target DNA from H. zea Transib, retrovirus integrases, Mos1 transposase and Mu transposase STC structures. Target site DNAs are shown as green and red. The approximate degree of bending in each target DNA is indicated. H. zea Transib is the only DDE/D-family transposase–integrase for which a STC structure has been reported that lacks a bend or base-unpairing at the centre of the target site DNA. Instead, Transib strongly bends target DNA near both edges of the target site DNA (between position −2 and −1 and positions 1 and 2), leading to a total 150° directional change of target DNA. Target DNAs in retroviral integrase STC structures exhibit relatively mild bends with one backbone kink at the centre of target site DNA, regardless of its length (ranging from 4 bp in PFV integrase to 6 bp in RSV integrase). The sharp bending (about 150°) at the centre of the Mos1 target DNA is achieved by flipping of the adenines in the TA target site. The target DNA in Mu STC exhibits a more continuous bending pattern through the 5-bp target site DNA, with one bend before the target site (between position −3 and −2), one at the centre and one immediately after the target site DNA (between position 2 and 3). The central bend is facilitated by the T–T mismatch in the target site. d, Transposon end–target DNA junction region of the Transib STC model superimposed on the cryo-EM map contoured at 5.5σ. Nucleotide residues in target DNA are labelled with a subscript T. e, Difference density between the Transib STC cryo-EM map and the model, showing the uncleaved target DNA phosphodiester bond in a portion of the particles used for cryo-EM map reconstruction. The difference map was contoured at 6σ. f, Superimposition of Transib TCC (protein in orange and metal ions in green) active site with Transib HFC active site (protein in purple and metal ions in grey). Distances are expressed in Å. Attacking oxygen atoms in HFC and TCC are highlighted in black and red circles, respectively. In TCC, the phosphorus is 2.4 Å from the attacking oxygen and the two metal ions are 3.2 Å apart. These distances are 3.6 Å and 4.2 Å in HFC. g, Sequence alignment of Transib transposases, vertebrate RAG1 and deuterostome invertebrate RAG1L proteins, showing the regions corresponding to three RAG2-binding interfaces in RAG1. Residue numbers are for H. zea Transib. Hs, Homo sapiens (human); Mm, Mus musculus (mouse); Dr, Danio rerio (zebrafish); Gg, Gallus gallus (chicken); Bb, Branchiostoma belcheri (amphioxus); Sp, Strongylocentrotus purpuratus (purple sea urchin); Pf, Ptychodera flava (acorn worm); Pm, Petromyzon marinus (sea lamprey) and Af, Asterias forbesi (sea star).
Extended Data Fig. 8 Structural insights into the function and evolution of H. zea Transib CTT.
a, Interactions between Transib CTT α20 and ZnB domain α12–α13. Residues in CTT and ZnB are coloured red and green, respectively. Residues involved in hydrophobic interactions are shown in ball-and-stick representation. b, Superimposition of ZnB domain (pale colours) together with CTT α20 (dark colours) from the structures representing five steps in transposition. c, Cleavage of DNA substrates bearing a 5′TIR–3′TIR pair by MBP-tagged wild-type or CTT truncated mutant Transib transposases, each with N-terminal 16 amino acids removed. The DNA cleavage products were resolved on a 6% TBE polyacrylamide gel and stained with SYBR Gold. Open and closed arrowheads indicate single 5′TIR and single 3′TIR cleavage products, respectively. The red asterisk marks the double-cleavage band. The experiment was repeated at least three times independently and similar results were obtained. For gel source data, see Supplementary Fig. 1. d, Superimposition of Transib, RAG1 and BbRAG1L structures by the first two helices of their CTDs. Transib and BbRAG1L CTT extend from the structurally conserved CTD and point in different directions. e, Sequence alignment of H. zea Transib CTT with vertebrate RAG1 CTT and deuterostome invertebrate RAG1L CTT showing highly divergent sequences among the three groups. Residues mediating the hydrophobic interactions between ZnB α12–α13 and CTT α20 are highlighted in green. Residue numbers and secondary structure elements at the top of the sequence alignment are for H. zea Transib. The residue number for the final amino acid in the sequence alignment is indicated for selected sequences.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1: Raw, uncropped gel images for results obtained by gel-based electrophoretic separation and Supplementary Table 1: The sequences of target site duplications (TSDs) generated by HzTransib mediated transposition.
Video 1
A composite morph of two cycles of conformational changes of HzTransib during transposition. One ZnB domain is in green and one CTT domain is in red. Names of the structures and key events at each step are indicated.
Rights and permissions
About this article
Cite this article
Liu, C., Yang, Y. & Schatz, D.G. Structures of a RAG-like transposase during cut-and-paste transposition. Nature 575, 540–544 (2019). https://doi.org/10.1038/s41586-019-1753-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1753-7
This article is cited by
-
Zinc-finger BED domains drive the formation of the active Hermes transpososome by asymmetric DNA binding
Nature Communications (2023)
-
From parasites to partners: exploring the intricacies of host-transposon dynamics and coevolution
Functional & Integrative Genomics (2023)
-
Structural insight into Tn3 family transposition mechanism
Nature Communications (2022)
-
Structural insights into the evolution of the RAG recombinase
Nature Reviews Immunology (2022)
-
Identification of RAG-like transposons in protostomes suggests their ancient bilaterian origin
Mobile DNA (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.