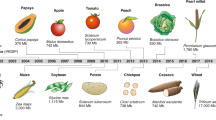
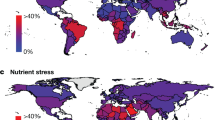
Abstract
Crop production needs to increase to secure future food supplies, while reducing its impact on ecosystems. Detailed characterization of plant genomes and genetic diversity is crucial for meeting these challenges. Advances in genome sequencing and assembly are being used to access the large and complex genomes of crops and their wild relatives. These have helped to identify a wide spectrum of genetic variation and permitted the association of genetic diversity with diverse agronomic phenotypes. In combination with improved and automated phenotyping assays and functional genomic studies, genomics is providing new foundations for crop-breeding systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Drèze, J. & Sen, A. K. Hunger and Public Action (Clarendon, 1989).
Foley, J. A. et al. Solutions for a cultivated planet. Nature 478, 337–342 (2011).
Kitano, M. et al. Ammonia synthesis using a stable electride as an electron donor and reversible hydrogen store. Nature Chem. 4, 934–940 (2012).
Zhao, C. et al. Plausible rice yield losses under future climate warming. Nature Plants 3, 16202 (2016).
Garrett, K. A., Dendy, S. P., Frank, E. E., Rouse, M. N. & Travers, S. E. Climate change effects on plant disease: genomes to ecosystems. Annu. Rev. Phytopathol. 44, 489–509 (2006).
Godfray, H. C. J. et al. Food security: the challenge of feeding 9 billion people. Science 327, 812–818 (2010).
Tilman, D., Balzer, C., Hill, J. & Befort, B. L. Global food demand and the sustainable intensification of agriculture. Proc. Natl Acad. Sci. USA 108, 20260–20264 (2011).
Bennett, M. D. & Leitch, I. J. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann. Bot. 107, 467–590 (2011).
Bennetzen, J. L., Ma, J. & Devos, K. M. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 95, 127–132 (2005).
Lisch, D. How important are transposons for plant evolution? Nature Rev. Genet. 14, 49–61 (2012).
Kim, M. Y. & Zilberman, D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 19, 320–326 (2014).
Jiao, Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2012).
Woodhouse, M. R. et al. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homeologs. PLoS Biol. 8, e1000409 (2010).
Neale, D. B. et al. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15, R59 (2014).
Shulaev, V. et al. The genome of woodland strawberry (Fragaria vesca). Nature Genet. 43, 109–116 (2010).
Chalhoub, B. et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014).
Bertioli, D. J. et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nature Genet. 48, 438–446 (2016).
Voskoboynik, A. et al. The genome sequence of the colonial chordate, Botryllus schlosseri . eLife 2, e00569 (2013).
Safar, J. et al. Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J. 39, 960–968 (2004).
International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788 (2014).
Sierro, N. et al. The tobacco genome sequence and its comparison with those of tomato and potato. Nature Commun. 5, 3833 (2014).
Zhang, T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnol. 33, 531–537 (2015).
Staňková, H. et al. BioNano genome mapping of individual chromosomes supports physical mapping and sequence assembly in complex plant genomes. Plant Biotechnol. J. 14, 1523–1531 (2016).
Yang, J. et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nature Genet. 48, 1225–1232 (2016).
Chaisson, M. J. P., Wilson, R. K. & Eichler, E. E. Genetic variation and the de novo assembly of human genomes. Nature Rev. Genet. 16, 627–640 (2015).
Kozarewa, I. et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nature Methods 6, 291–295 (2009).
Weisenfeld, N. I. et al. Comprehensive variation discovery in single human genomes. Nature Genet. 46, 1350–1355 (2014). This paper highlights the development and application of the DISCOVAR assembler, which has been of crucial importance for the creation of assemblies with improved representation of sequence variants.
Love, R. R., Weisenfeld, N. I., Jaffe, D. B., Besansky, N. J. & Neafsey, D. E. Evaluation of DISCOVAR de novo using a mosquito sample for cost-effective short-read genome assembly. BMC Genomics 17, 187 (2016).
Pendleton, M. et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nature Methods 12, 780–786 (2015). This article shows how the application of hybrid assembly methods has set new standards for sequence contiguity and the representation of diversity.
Zimin, A. et al. Sequencing and assembly of the 22-Gb loblolly pine genome. Genetics 196, 875–890 (2014).
Zimin, A. V. et al. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the mega-reads algorithm. Genome Res. http://dx.doi.org/10.1101/gr.213405.116 (2017). This paper shows how long-read sequencing technology coupled with the mega-reads algorithm can be used successfully to tackle a large and complex grass genome, which paves the way for the sequencing of multiple variants.
Chin, C.-S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nature Methods 13, 1050–1054 (2016). This study applies the PacBio long-read sequencing technology to resolving the highly heterozygous Vitis vinifera cv. Cabernet Sauvignon genome and demonstrates the importance of this technology for the assembly of complex plant genomes.
Koren, S. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nature Biotechnol. 30, 693–700 (2012).
Goodwin, S. et al. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 25, 1750–1756 (2015).
Loman, N. J., Quick, J. & Simpson, J. T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nature Methods 12, 733–735 (2015). Refs 34 and 35 highlight the potential of nanopore sequencing technology, using yeast and bacterial genomes.
Burton, J. N. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nature Biotechnol. 31, 1119–1125 (2013).
Session, A. M. et al. Genome evolution in the allotetraploid frog Xenopus laevis . Nature 538, 336–343 (2016).
Selvaraj, S., Dixon, J. R., Bansal, V. & Ren, B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nature Biotechnol. 31, 1111–1118 (2013).
Putnam, N. H. et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26, 342–350 (2016).
Jarvis, D. E. et al. The genome of Chenopodium quinoa . Nature 542, 307–312 (2017).
Mostovoy, Y. et al. A hybrid approach for de novo human genome sequence assembly and phasing. Nature Methods 13, 587–590 (2016).
Zheng, G. X. Y. et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nature Biotechnol. 34, 303–311 (2016). As well as refs 41 and 43, this paper shows the considerable potential of linked-read sequencing technology for resolving the phasing of complete chromosomes.
Weisenfeld, N. I., Kumar, V., Shah, P., Church, D. & Jaffe, D. B. Direct determination of diploid genome sequences. Preprint at http://biorxiv.org/content/early/2016/08/19/070425 (2016).
Denoeud, F. et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345, 1181–1184 (2014).
D'Hont, A. et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217 (2012).
Clavijo, B. J. et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Preprint at http://biorxiv.org/content/early/2016/11/04/080796 (2016). This preprint presents open-source assembly methods that preserve genetic variation and have enabled the fast and low-cost assembly of the large and complex wheat genome.
Grivet, L. & Arruda, P. Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr. Opin. Plant Biol. 5, 122–127 (2001).
Byrne, S. L. et al. A synteny-based draft genome sequence of the forage grass Lolium perenne . Plant J. 84, 816–826 (2015).
Bilgic, H., Hakki, E. E., Pandey, A., Khan, M. K. & Akkaya, M. S. Ancient DNA from 8400 year-old Çatalhöyük wheat: implications for the origin of Neolithic agriculture. PLoS ONE 11, e0151974 (2016).
Dvorak, J., Luo, M.-C. & Akhunov, E. D. N. I. Vavilov's theory of centres of diversity in the light of current understanding of wheat diversity, domestication and evolution. Czech J. Genet. Plant Breed. 47, S20–S27 (2011).
Zheng, Y., Crawford, G. W., Jiang, L. & Chen, X. Rice domestication revealed by reduced shattering of archaeological rice from the lower Yangtze valley. Sci. Rep. 6, 28136 (2016).
Huang, X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012).
Doebley, J. F., Gaut, B. S. & Smith, B. D. The molecular genetics of crop domestication. Cell 127, 1309–1321 (2006).
Buckler, E. S. et al. The genetic architecture of maize flowering time. Science 325, 714–718 (2009).
Ramos-Madrigal, J. et al. Genome sequence of a 5,310-year-old maize cob provides insights into the early stages of maize domestication. Curr. Biol. 26, 3195–3201 (2016).
Wright, S. I. et al. The effects of artificial selection on the maize genome. Science 308, 1310–1314 (2005).
Galili, G., Levy, A. A. & Feldman, M. Gene-dosage compensation of endosperm proteins in hexaploid wheat Triticum aestivum . Proc. Natl Acad. Sci. USA 83, 6524–6528 (1986).
Zhang, Z. et al. Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc. Natl Acad. Sci. USA 108, 18737–18742 (2011).
Dubcovsky, J. & Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866 (2007).
Khoury, C. K. et al. Increasing homogeneity in global food supplies and the implications for food security. Proc. Natl Acad. Sci. USA 111, 4001–4006 (2014).
Massawe, F., Mayes, S. & Cheng, A. Crop diversity: an unexploited treasure trove for food security. Trends Plant Sci. 21, 365–368 (2016).
Tanksley, S. D. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066 (1997).
Jafarzadeh, J. et al. Breeding value of primary synthetic wheat genotypes for grain yield. PLoS ONE 11, e0162860 (2016).
Munns, R. et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnol. 30, 360–364 (2012).
Borrill, P., Adamski, N. & Uauy, C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 208, 1008–1022 (2015).
McCouch, S. R. et al. Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa . Euphytica 154, 317–339 (2006).
Liu, Z. et al. Expanding maize genetic resources with predomestication alleles: maize–teosinte introgression populations. Plant Genome http://dx.doi.org/10.3835/plantgenome2015.07.0053 (2016).
Witek, K. et al. Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nature Biotechnol. 34, 656–660 (2016).
Steuernagel, B. et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nature Biotechnol. 34, 652–655 (2016). This paper highlights a method with great promise for capturing the diversity of large genes families in populations of crops and their wild relatives.
Krasileva, K. V. et al. Uncovering hidden variation in polyploid wheat genomes. Proc. Natl Acad. Sci. USA 114, E913–E921 (2017). This paper describes functional genome resources that have been developed for tetraploid and hexaploid wheat lines — resources that will expedite many new areas of research.
Zhang, Y. et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nature Commun. 7, 12617 (2016).
Gil-Humanes, J. et al. High efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. http://dx.doi.org/10.1111/tpj.13446 (2016).
Biffen, R. H. & Engledow, F. L. Wheat-Breeding Investigations at the Plant Breeding Institute, Cambridge (His Majesty's Stationery Office, 1926).
Allen, A. M. et al. Characterization of a Wheat Breeders' Array suitable for high throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivium). Plant Biotechnol. J. http://dx.doi.org/10.1111/pbi.12635 (2016).
Barabaschi, D. et al. Next generation breeding. Plant Sci. 242, 3–13 (2015).
Bassi, F. M., Bentley, A. R., Charmet, G., Ortiz, R. & Crossa, J. Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci. 242, 23–36 (2016).
Vivek, B. S. et al. Use of genomic estimated breeding values results in rapid genetic gains for drought tolerance in maize. Plant Genome http://dx.doi.org/10.3835/plantgenome2016.07.0070 (2017).
Riaz, A., Periyannan, S., Aitken, E. & Hickey, L. A rapid phenotyping method for adult plant resistance to leaf rust in wheat. Plant Methods 12, 17 (2016).
Varshney, R. K., Terauchi, R. & McCouch, S. R. Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol. 12, e1001883 (2014).
Rodgers-Melnick, E., Vera, D. L., Bass, H. W. & Buckler, E. S. Open chromatin reveals the functional maize genome. Proc. Natl Acad. Sci. USA 113, E3177–E3184 (2016).
Marulanda, J. J. et al. Optimum breeding strategies using genomic selection for hybrid breeding in wheat, maize, rye, barley, rice and triticale. Theor. Appl. Genet. 129, 1901–1913 (2016).
Spindel, J. E. et al. Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity 116, 395–408 (2016).
Patil, G. et al. Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci. Rep. 6, 19199 (2016).
Jordan, K. W., Wang, S., Lun, Y. & Gardiner, L. J. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol. 16, 48 (2015).
Porreca, G. J. et al. Multiplex amplification of large sets of human exons. Nature Methods 4, 931–936 (2007).
Saintenac, C. et al. Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.). Genetics 181, 393–403 (2008).
Sadhu, M. J., Bloom, J. S., Day, L. & Kruglyak, L. CRISPR-directed mitotic recombination enables genetic mapping without crosses. Science 352, 1113–1116 (2016).
Furbank, R. T. & Tester, M. Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 16, 635–644 (2011).
Fiorani, F. & Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 64, 267–291 (2013).
Araus, J. L. & Cairns, J. E. Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 19, 52–61 (2013).
Zamir, D. Where have all the crop phenotypes gone? PLoS Biol. 11, e1001595 (2013).
Loose, M., Malla, S. & Stout, M. Real-time selective sequencing using nanopore technology. Nature Methods 13, 751–754 (2016).
Rand, A. C. et al. Cytosine variant calling with high-throughput nanopore sequencing. Preprint at http://biorxiv.org/content/early/2016/04/04/047134 (2016).
Simpson, J. T. et al. Detecting DNA methylation using the Oxford Nanopore Technologies MinION sequencer. Preprint at http://biorxiv.org/content/early/2016/04/04/047142 (2016).
Acknowledgements
This work was supported by strategic programme funding from the UK Biotechnology and Biological Sciences Research Council (BBSRC) (GRO BB/J004588/1) to M.B., a BBSRC strategic LoLa award (BB/J003913/1) to M.B. and M.C. and funding from the Gatsby Charitable Foundation to K.K. and the 2Blades Foundation to B.W.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Reviewer Information Nature thanks V. Albert, J. Schmutz, T. Mitchell-Olds and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Rights and permissions
About this article
Cite this article
Bevan, M., Uauy, C., Wulff, B. et al. Genomic innovation for crop improvement. Nature 543, 346–354 (2017). https://doi.org/10.1038/nature22011
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature22011
This article is cited by
-
Integrated meta-analysis and transcriptomics pinpoint genomic loci and novel candidate genes associated with submergence tolerance in rice
BMC Genomics (2024)
-
Innovative computational tools provide new insights into the polyploid wheat genome
aBIOTECH (2024)
-
Male sterility systems and their applications in hybrid wheat breeding
Cereal Research Communications (2024)
-
Pan-3D genome analysis reveals structural and functional differentiation of soybean genomes
Genome Biology (2023)
-
Lifting of the 1,000 wheat exome project SNPs from Triticum aestivum cv. Chinese Spring assembly RefSeq v1.0 to RefSeq v2.1
BMC Research Notes (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.