Abstract
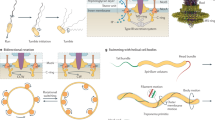
Various rod-shaped bacteria mysteriously glide on surfaces in the absence of appendages such as flagella or pili. In the deltaproteobacterium Myxococcus xanthus, a putative gliding motility machinery (the Agl–Glt complex) localizes to so-called focal adhesion sites (FASs) that form stationary contact points with the underlying surface. Here we show that the Agl–Glt machinery contains an inner-membrane motor complex that moves intracellularly along a right-handed helical path; when the machinery becomes stationary at FASs, the motor complex powers a left-handed rotation of the cell around its long axis. At FASs, force transmission requires cyclic interactions between the molecular motor and the adhesion proteins of the outer membrane via a periplasmic interaction platform, which presumably involves contractile activity of motor components and possible interactions with peptidoglycan. Our results provide a molecular model of bacterial gliding motility.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Islam, S. T. & Mignot, T. The mysterious nature of bacterial surface (gliding) motility: A focal adhesion-based mechanism in Myxococcus xanthus. Semin. Cell Dev. Biol. 46, 143–154 (2015)
Luciano, J. et al. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet. 7, e1002268 (2011)
Sun, M., Wartel, M., Cascales, E., Shaevitz, J. W. & Mignot, T. Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl Acad. Sci. USA 108, 7559–7564 (2011)
Jakobczak, B., Keilberg, D., Wuichet, K. & Søgaard-Andersen, L. Contact- and protein transfer-dependent stimulation of assembly of the gliding motility machinery in Myxococcus xanthus. PLoS Genet. 11, e1005341 (2015)
Nan, B. et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc. Natl Acad. Sci. USA 108, 2498–2503 (2011)
Balagam, R. et al. Myxococcus xanthus gliding motors are elastically coupled to the substrate as predicted by the focal adhesion model of gliding motility. PLOS Comput. Biol. 10, e1003619 (2014)
Treuner-Lange, A. et al. The small G-protein MglA connects to the MreB actin cytoskeleton at bacterial focal adhesions. J. Cell Biol. 210, 243–256 (2015)
Nan, B. et al. Flagella stator homologs function as motors for myxobacterial gliding motility by moving in helical trajectories. Proc. Natl Acad. Sci. USA 110, E1508–E1513 (2013)
Kaiser, D. & Warrick, H. Transmission of a signal that synchronizes cell movements in swarms of Myxococcus xanthus. Proc. Natl Acad. Sci. USA 111, 13105–13110 (2014)
Bean, G. J. et al. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry 48, 4852–4857 (2009)
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008)
Wojdyla, J. A. et al. Structure and function of the Escherichia coli Tol-Pal stator protein TolR. J. Biol. Chem. 290, 26675–26687 (2015)
Gresock, M. G., Kastead, K. A. & Postle, K. From homodimer to heterodimer and back: elucidating the TonB energy transduction cycle. J. Bacteriol. 197, 3433–3445 (2015)
Wartel, M. et al. A versatile class of cell surface directional motors gives rise to gliding motility and sporulation in Myxococcus xanthus. PLoS Biol. 11, e1001728 (2013)
Bui, N. K. et al. The peptidoglycan sacculus of Myxococcus xanthus has unusual structural features and is degraded during glycerol-induced myxospore development. J. Bacteriol. 191, 494–505 (2009)
Cascales, E., Gavioli, M., Sturgis, J. N. & Lloubès, R. Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli. Mol. Microbiol. 38, 904–915 (2000)
Wang, S., Furchtgott, L., Huang, K. C. & Shaevitz, J. W. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc. Natl Acad. Sci. USA 109, E595–E604 (2012)
Ejigiri, I. et al. Shedding of TRAP by a rhomboid protease from the malaria sporozoite surface is essential for gliding motility and sporozoite infectivity. PLoS Pathog. 8, e1002725 (2012)
den Blaauwen, T., de Pedro, M. A., Nguyen-Distèche, M. & Ayala, J. A. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32, 321–344 (2008)
Nan, B., Mauriello, E. M. F., Sun, I.-H., Wong, A. & Zusman, D. R. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol. Microbiol. 76, 1539–1554 (2010)
Bustamante, V. H., Martínez-Flores, I., Vlamakis, H. C. & Zusman, D. R. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol. Microbiol. 53, 1501–1513 (2004)
Ducret, A., Théodoly, O. & Mignot, T. Single cell microfluidic studies of bacterial motility. Methods Mol. Biol. 966, 97–107 (2013)
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997)
Johnson, L. S., Eddy, S. R. & Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11, 431 (2010)
Söding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005)
Holm, L., Kääriäinen, S., Rosenström, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008)
Lassmann, T., Frings, O. & Sonnhammer, E. L. L. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 37, 858–865 (2009)
Pei, J., Sadreyev, R. & Grishin, N. V. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics 19, 427–428 (2003)
Drozdetskiy, A., Cole, C., Procter, J. & Barton, G. J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43 (W1), W389–W394 (2015)
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014)
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001)
Käll, L., Krogh, A. & Sonnhammer, E. L. L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 (2004)
Marbouty, M. et al. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell 59, 588–602 (2015)
Ducret, A., Valignat, M. P., Mouhamar, F., Mignot, T. & Theodoly, O. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc. Natl Acad. Sci. USA 109, 10036–10041 (2012)
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012)
Arigovindan, M. et al. High-resolution restoration of 3D structures from widefield images with extreme low signal-to-noise-ratio. Proc. Natl Acad. Sci. USA 110, 17344–17349 (2013)
Marques, O. Practical Image and Video Processing using MATLAB (John Wiley & Sons, 2011)
Wolter, S. et al. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods 9, 1040–1041 (2012)
Acknowledgements
We thank R. Mercier for suggestions and critical reading of the manuscript, A. Valeri for help with PCA analysis, C. Fiche for help with image analysis and Y. Denis (IMM transcriptomics platform) for help with quantitative PCR and M. Wartel for preliminary experiments. L.M.F. was partially funded by Fondation ARC, L.E. was supported by the Fondation pour la Recherche Médicale (FRM DGE2010221257), S.T.I. was supported by a fellowship from the Canadian Institutes of Health Research and the AMIDEX program of Aix-Marseille Université, and A.L. and V.A. were supported by the IRP funds of the NIH, NIH grant GM113172 to M.S.V.N. and Y.V.B. and NIH grant GM51986 to Y.V.B. Research in the Mignot laboratory was supported by European Research Council grant DOME-261105 and a Bettencourt-Schueller “Coup d’élan pour la recherche Française 2011”. Research in the Nollmann laboratory was supported by ANR grants IBM (ANR-14-CE09-0025-01), HiResBacs (ANR-15-CE11-0023), and European Research Council grant smInsulator-260787. We acknowledge support from France-BioImaging (ANR-10-INBS-04, “Investments for the future”), and Imagine Optic.
Author information
Authors and Affiliations
Contributions
L.M.F., J.-B.F., L.E., M.N. and T.M. conceived the experiments and analysed the data with help from A.D. for TADA experiment analysis. L.M.F. and J.-B.F. performed most of the experiments, including TIRF assays, astigmatism and strain construction. A.D. and S.L. helped with TADA experiments. J.L., M.S., and S.L. constructed strains. S.T.I., J.T. and O.T. performed RICM studies. V.A. and A.L. performed bioinformatics analysis. E.K., M.S.V.N. and Y.V.B. provided TADA. M.N. and T.M. wrote the paper with help from L.M.F., J.-B.F. and L.E.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks J. Armitage, P. Kukura and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Dynamic AglZ–YFP clusters in non-motile cells.
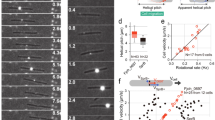
a, Dynamic AglZ–YFP clusters in a non-motile cell observed by TIRFM. Kymograph representation of cluster movement captured every 0.5 s. Note that the clusters form at the cell pole and move directionally towards the opposite cell pole where they are dispersed. Scale bar, 2 μm. b, Distribution of cluster speeds between and within cells. Note that clusters can move at different speeds in a cell and that the speed between cells generally varies between 2 and 4 μm min−1 (two technical replicates). c, Number of AglZ–YFP clusters per cell per minute in wild-type and A22-treated cells (two technical replicates). d, AglZ–YFP cluster speed in wild-type and A22-treated cells (two technical replicates). e, Trajectory angles in wild-type and A22-treated cells (two technical replicates). Wilcoxon tests; *P < 0.1; **P < 0.01; ***P < 0.001 (c–e).
Extended Data Figure 2 Myxococcus cells rotate along their long axes during motility.
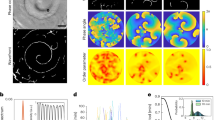
a, TADA-bright clusters form in Myxococcus cells subjected to a brief osmotic shock. TADA is incorporated in the Myxococcus cell envelope only when the cells are subjected to NaCl treatment (see Methods). b, c, TADA-bright clusters are not dynamic in non-motile cells. TADA-bright cluster movements are not detectable in non-motile cells (c, red dots, n = 14; two technical replicates) and only detectable in moving cells (c, blue dots, n = 17; two technical replicates). MSD, mean square displacement. Scale bar, 2 μm. d, TADA-bright clusters are inert. FRAP analysis reveals the absence of fluorescent molecule exchange in TADA-bright clusters (n = 6; one technical experiment). Scale bar, 2 μm. e, TADA-bright cluster rotation reflects rotation of the cell during movement. A cell on which four TADA-bright clusters were tracked is shown. The radial velocity of each cluster calculated by projection of the 2D images on the model 3D cell cylinder (left, β angle) is plotted against the linear displacement of the cell. Each TADA cluster moved at the same radial speed and proportionally to the speed of the cell, indicating that TADA clusters are inert objects reporting on the rigid-body movement of the cell. f, Three dimensional trajectories of TADA-bright clusters reconstructed by astigmatism. In the absence of astigmatism, the size of TADA clusters corresponds to the diffraction limit of light, and they are circular (that is, the size of the PSF is the same in perpendicular directions), making the astigmatic analysis of axial position possible. g, TADA-bright clusters rotate clockwise. Cluster intensity fluctuations and positions relative to the cell axis are shown over time (left) in a representative cell (right). The black arrow points to the analysed cluster. The representative cell shown in the right panel was isolated from others in the field with a black mask (n = 10; three technical replicates).
Extended Data Figure 3 Analysis of AglZ–YFP, GltI–YFP and AglQ–mCherry in agl and glt mutant backgrounds.
a, Each fluorescent functional fusion gene was introduced in place of the wild-type gene in each genetic background shown. Typical examples are shown for each strain. Crossed boxes indicated genetic backgrounds that were not obtained for this study. Scale bar, 2 μm. b, Cluster detection and analysis chart. Phase contrast and fluorescence images were processed so as to respectively extract cell masks of isolated cells (compared edited mask to raw mask) and the position of fluorescence clusters following the application of a fluorescence bandpass filter. Note that the intensity of the fluorescence clusters was not exploited owing to lack of robustness and day-to-day fluctuations. The cluster coordinates were then defined relative to cell coordinates with the Microbe J plugin (http://www.indiana.edu/~microbej/) in Fiji, compiled in R sheets and further analysed by PCA using custom-written code in Matlab.
Extended Data Figure 4 PCA of AglZ–YFP, GltI–YFP and AglQ–mCherry in agl and glt mutant backgrounds.
a–c, Coefficients of the principal components (PCs) for AglZ–YFP, GltI–YFP and AglQ–mCherry. PCs are the eigenvectors of the correlation matrix calculated from the four parameters indicated in the first column of the tables. Together, PCs form an orthogonal basis in which the vectors are uncorrelated. PCs are sorted according to the amount of variability in the data they describe, PC1 having the largest effect (that is, variance) and PC4 the least. d, Scree plots displaying the variance associated to each PC. The bar plot represents the variance associated with each PC for a given fusion (from left to right: AglZ–YFP, GltI–YFP and AglQ–mCherry). The cumulative variance is also plotted (light blue line). Note that PC1–2 describe on average 70% of the total variance and PC1–3 more than 87%. e, Projection of the data in the space defined by the three first PCs. For each mutant, data are represented by a scatter plot of a specific colour (see inset for colour code). For each direction and each mutant, the average and s.d. of the data are symbolized by a single bold line: the centre of the line represents the average and its length the s.d.
Extended Data Figure 5 Motility is propelled by cyclic interactions between the inner membrane-localized motor and outer membrane-localized adhesins of the motility complex.
a, Epifluorescence analysis of representative cells expressing AglZ–YFP–AglQ/GltD/GltC–mCherry. In each case, fluorescent functional fusions are expressed in place of the wild-type gene. Note that while AglZ–YFP and AglQ–mCherry clusters can be detected, GltC–mCherry and GltD–mCherry appear mostly diffuse around the cell envelope with these imaging conditions. Forty cells were imaged for AglZ–YFP–AglQ with two biological replicates; 232 cells were imaged for AglZ–YFP–GltD with three biological replicates; and 55 cells were imaged for AglZ–YFP–GltC with three biological replicates. Scale bar, 2 μm. b, Protein co-localization in static clusters. For each cell analysed, a percentage of co-localization is computed for proteins detected only in static clusters. Values range from 0 when no co-localization between the two proteins was detected in the cell to 100% when the two proteins were always detected together. The left panel shows the percentage of co-localization for AglQ, GltD or GltI with respect to AglZ. Single data are represented by a scatter plot (o), the median co-localization value is symbolized by a blue line and the s.d. by light grey boxes. On average, 96% of AglQ clusters colocalized with AglZ (n = 153 clusters; two biological replicates), 96% of GltD (n = 120; seven biological replicates) and 83% for GltC (n = 100; three biological replicates). Inversely, the right panel represents the percentage of co-localization of AglZ with respect to AglQ (97%, n = 152; two biological replicates), GltD (91%, n = 120; seven biological replicates) and GltC (69%, n = 100; three biological replicates), respectively. c, Protein co-localization in mobile clusters. Box-plots read as in b and describe the percentage of co-localization for proteins detected only in dynamic clusters. The left panel illustrates the co-localization of AglQ, GltD or GltC with respect to AglZ (76%, n = 106, two biological replicates; 39%, n = 11, seven biological replicates; and 0%, n = 4, three biological replicates, respectively). The right panel shows the percentage of co-localization of AglZ with respect to AglQ (41%, n = 125; two biological replicates), GltD (0%, n = 72; seven biological replicates) and GltC (0%, n = 41; three biological replicates). From the two panels, it appears that co-localization in dynamic clusters is essentially observed between AglZ and AglQ. For GltD and GltC, dynamic clusters are almost never detected (n = 11 for GltD, n = 4 for GltC) leading to a co-localization score close or equal to 0%. d, AglZ–YFP clusters localize within adhesive contact zones. Left, RICM of a representative gliding cell (n = 10, two biological replicates, 30-s time frames; scale bar, 2 μm) showing intimate connection with the chitosan-coated glass surface (dark zone). Right, adhesions and AglZ–YFP cluster localization in detaching cells by RICM and combined epifluorescence microscopy (time frames, 30 s; scale bar, 2 μm). The graph represents the distribution of RICM intensities at AglZ–YFP cluster positions (red line) compared to the average intensity along the whole cell body (black line). Data obtained for n = 20 cells; two biological replicates. e, Gliding Myxococcus cells deposit outer membrane vesicles in their wake. TIRFM images of a motile cell expressing both outer membrane sfGFP and inner membrane mCherry. Outer membrane vesicles are deposited, suggesting that the cell is firmly adhered to the underlying surface. Shown is a representative cell (n = 60; 12 technical replicates). f, GltD–mCherry, a periplasmic Glt protein, is not released by gliding cells at focal adhesion complexes. Top, TIRFM snapshots of a representative cell expressing both GltD–mCherry (red) and AglZ–YFP (green). The position of the GltD clusters on the surface coincides with position of focal adhesion complexes (white asterisk). Time frames, 15 s; scale bar, 2 μm. Bottom, variation of intensity for GltD–mcherry (red) and AglZ–YFP (green) as a function of time before (negative time) and after (positive time) the cell moved away from the focal adhesion position, shown for n = 10 cells (two biological replicates).
Extended Data Figure 6 Image analysis for TIRFM experiments.
a, Temporal RGB image computed from fluorescence images (mCherry for this example). Images were summed together and colour coded from blue (first image) to red (last image). Immobile cells appear uniformly white whereas moving cells show coloured extremities. Cell trajectory is represented by a yellow dotted line. b, For the two imaging channels (YFP and mCherry), a kymograph and a montage were calculated. Kymograph reads from top (first image) to bottom (last image), each line representing the average fluorescent intensity computed along the cell trajectory. The montage shows for each acquisition an image of the cell after applying a straightening algorithm. Clusters of proteins (AglZ–YFP or AglQ/GltD/GltI–mCherry) appear as bright spots at the centre of the cell. c, Kymographs after applying a denoising algorithm. The cell outline was depicted either by a dotted white line when the cell was immobile or by a continuous white line when the cell was gliding on the surface. d, Cell speed as a function of acquisition time. For each cell, a threshold was defined that depended on the SNR and the sample lateral stability during TIRFM acquisition. When the cell speed was below this threshold (horizontal dotted line), the cell was considered immobile. Phases associated with cell movement are highlighted in blue.
Extended Data Figure 7 Co-localization estimation and correlation with cell movement.
a, Co-localization between YFP-tagged and mCherry-tagged proteins was calculated in three steps. For each channel (YFP and mCherry), a threshold was applied to the montage in order to detect protein clusters (clusterization). When the same cluster was observed in successive images, its localizations were stitched together manually and finally classified as either static (red) or dynamic/mobile (blue). b, For each channel, a protein-detection map was computed from the kymograph and the positions of the detected clusters. The cell outline was depicted by two white lines. c, The co-localization map was obtained by multiplying the two protein-detection maps. In the inset, the cumulative intensities associated with static (red) and dynamic (blue) clusters are plotted as a function of time. d, Cell speed as a function of acquisition time. Blue boxes represent regions in which cell speed is higher than the threshold. e, Cumulative co-localization intensity (static and dynamic) as a function of acquisition time. Blue boxes represent regions with high cell speeds. f, Curves from d and e were binarized. For the cell speed, the value was set to ‘0’ when the speed was below the threshold (immobile cell) and to ‘1’ when it was above (gliding cell) (left). For the co-localization, the value was set to ‘1’ when co-localization was detected, ‘0’ otherwise (left). A correlation curve (right) was then computed by comparing the two curves. At each time-point, if the values of the two binarized curves were equal (1/1 or 0/0, green highlighted areas), the correlation was set to ‘1’. Otherwise, it was set to ‘−1’ (red highlighted area). Finally, the correlation score was defined as the average of all the correlation values.
Extended Data Figure 8 Bioinformatics analysis of AglQ/S and GltF.
a, AglQ and AglS carry a potential peptidoglycan (PG)-binding site. Multiple alignment of AglQ, AglS and their paralogues with TolR(5by4A) and ExbD (2pfuA). The gene name, organism name and GI (NCBI) accession number are given. The structure is shown on top and the 80% consensus is shown below the alignment. b, Cartoon views of the structures are shown below with the residues known or predicted to bind peptidoglycan shown as sticks. c, The GltF family of proteins found in deltaproteobacteria. Multiple alignment of the GltF family is shown and labelled using gene name, organism name and GI. The potential TonB box analogue is indicated. The TonB box can be most generally defined as an extended region that forms a β-strand-like structure that is not paired with other β-strands into a structural unit. The TonB box typically has two polar residues, T/S, and classically an acidic/amide residue. The GltF sequence profile analysis shows that GltF is related to the N-terminal region of certain OMP barrels that do contain a potential TonB-box like peptide.
Extended Data Figure 9 Bioinformatics analysis of the GltA/B/H system.
a, Multiple alignment of the β-barrel OMP proteins GltA, GltB and GltH and their paralogues with NspA (1p4tA). Note the presence of TonB box analogues in GltA and GltB but not GltH. b, Cartoon view of the inferred trimer of β-barrels based on the NspA structure with the residues predicted to form a trimeric interaction interface shown as spheres.
Supplementary information
Supplementary Table 1
Bioinformatic domain predictions in Agl-Glt proteins. (XLSX 11 kb)
Supplementary Table 2
Strains used in this study. (XLSX 10 kb)
Supplementary Table 3
Plasmids used in this study. (XLSX 9 kb)
Supplementary Table 4
Primers used in this study. (XLSX 8 kb)
Supplementary Table 5
Plasmid construction scheme. (XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Faure, L., Fiche, JB., Espinosa, L. et al. The mechanism of force transmission at bacterial focal adhesion complexes. Nature 539, 530–535 (2016). https://doi.org/10.1038/nature20121
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20121
This article is cited by
-
Filamentous structures in the cell envelope are associated with bacteroidetes gliding machinery
Communications Biology (2023)
-
Multi-scale dynamic imaging reveals that cooperative motility behaviors promote efficient predation in bacteria
Nature Communications (2023)
-
Bacterial motility: machinery and mechanisms
Nature Reviews Microbiology (2022)
-
Hidden paths to endless forms most wonderful: ecology latently shapes evolution of multicellular development in predatory bacteria
Communications Biology (2022)
-
1H, 13C and 15N chemical shift assignments of the ZnR and GYF cytoplasmic domains of the GltJ protein from Myxococcus xanthus
Biomolecular NMR Assignments (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.