Abstract
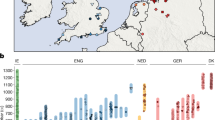
We generated genome-wide data from 69 Europeans who lived between 8,000–3,000 years ago by enriching ancient DNA libraries for a target set of almost 400,000 polymorphisms. Enrichment of these positions decreases the sequencing required for genome-wide ancient DNA analysis by a median of around 250-fold, allowing us to study an order of magnitude more individuals than previous studies1,2,3,4,5,6,7,8 and to obtain new insights about the past. We show that the populations of Western and Far Eastern Europe followed opposite trajectories between 8,000–5,000 years ago. At the beginning of the Neolithic period in Europe, ∼8,000–7,000 years ago, closely related groups of early farmers appeared in Germany, Hungary and Spain, different from indigenous hunter-gatherers, whereas Russia was inhabited by a distinctive population of hunter-gatherers with high affinity to a ∼24,000-year-old Siberian6. By ∼6,000–5,000 years ago, farmers throughout much of Europe had more hunter-gatherer ancestry than their predecessors, but in Russia, the Yamnaya steppe herders of this time were descended not only from the preceding eastern European hunter-gatherers, but also from a population of Near Eastern ancestry. Western and Eastern Europe came into contact ∼4,500 years ago, as the Late Neolithic Corded Ware people from Germany traced ∼75% of their ancestry to the Yamnaya, documenting a massive migration into the heartland of Europe from its eastern periphery. This steppe ancestry persisted in all sampled central Europeans until at least ∼3,000 years ago, and is ubiquitous in present-day Europeans. These results provide support for a steppe origin9 of at least some of the Indo-European languages of Europe.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
European Nucleotide Archive
Data deposits
The aligned sequences are available through the European Nucleotide Archive under accession number PRJEB8448. The Human Origins genotype dataset including ancient individuals can be found at (http://genetics.med.harvard.edu/reichlab/Reich_Lab/Datasets.html).
References
Fu, Q. et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014)
Gamba, C. et al. Genome flux and stasis in a five millennium transect of European prehistory. Nature Commun. 5, 5257 (2014)
Keller, A. et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nature Commun. 3, 698 (2012)
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014)
Olalde, I. et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228 (2014)
Raghavan, M. et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91 (2014)
Seguin-Orlando, A. et al. Genomic structure in Europeans dating back at least 36,200 years. Science 346, 1113–1118 (2014)
Skoglund, P. et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science 344, 747–750 (2014)
Anthony, D. W. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World (Princeton Univ. Press, 2007)
Fu, Q. et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl Acad. Sci. USA 110, 2223–2227 (2013)
Rohland, N., Harney, E., Mallick, S., Nordenfelt, S. & Reich, D. Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Phil. Trans. R. Soc. Lond. B 370, 20130624 (2015)
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012)
Fu, Q. et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 23, 553–559 (2013)
Brandt, G. et al. Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science 342, 257–261 (2013)
Der Sarkissian, C. et al. Ancient DNA reveals prehistoric gene-flow from Siberia in the complex human population history of North East Europe. PLoS Genet. 9, e1003296 (2013)
Briggs, A. W. et al. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38, e87 (2010)
Briggs, A. W. et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA 104, 14616–14621 (2007)
Myres, N. M. et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 19, 95–101 (2011)
Underhill, P. A. et al. The phylogenetic and geographic structure of Y-chromosome haplogroup R1a. Eur. J. Hum. Genet. 23, 124–131 (2015)
Skoglund, P. et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336, 466–469 (2012)
Czebreszuk, J. in Ancient Europe, 8000 B.C. to A.D. 1000: Encyclopedia of the Barbarian World (eds Bogucki, P. I. & Crabtree, P. J. ) 467–475 (Charles Scribners & Sons, 2003)
Lipson, M. et al. Efficient moment-based inference of admixture parameters and sources of gene flow. Mol. Biol. Evol. 30, 1788–1802 (2013)
Szécsényi-Nagy, A. et al. Tracing the genetic origin of Europe’s first farmers reveals insights into their social organization. Preprint at bioRxivhttp://dx.doi.org/10.1101/008664 (2014)
Haak, W. et al. Ancient DNA from European early Neolithic farmers reveals their Near Eastern affinities. PLoS Biol. 8, e1000536 (2010)
Hellenthal, G. et al. A genetic atlas of human admixture history. Science 343, 747–751 (2014)
Ralph, P. & Coop, G. The geography of recent genetic ancestry across Europe. PLoS Biol. 11, e1001555 (2013)
Renfrew, C. Archaeology and Language: The Puzzle of Indo-European Origins (Pimlico, 1987)
Bellwood, P. First Farmers: The Origins of Agricultural Societies (Wiley-Blackwell, 2004)
Gamkrelidze, T. V. & Ivanov, V. V. The early history of Indo-European languages. Sci. Am. 262, 110–116 (1990)
Mallory, J. P. In Search of the Indo-Europeans: Language, Archaeology and Myth (Thames and Hudson, 1991)
Kircher, M., Sawyer, S. & Meyer, M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2012)
Meyer, M. et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505, 403–406 (2014)
Rohland, N., Harney, E., Mallick, S., Nordenfelt, S. & Reich, D. Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Phil. Trans. R. Soc. Lond. B 370, 20130624 (2015)
Rohland, N. & Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946 (2012)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Behar, D. M. et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684 (2012)
Lassmann, T. & Sonnhammer, E. L. L. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6, 298 (2005)
Sawyer, S., Krause, J., Guschanski, K., Savolainen, V. & Pääbo, S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE 7, e34131 (2012)
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010)
Alexander, D. H. & Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12, 246 (2011)
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009)
Reich, D., Price, A. L. & Patterson, N. Principal component analysis of genetic data. Nature Genet. 40, 491–492 (2008)
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Skoglund, P., Storå, J., Götherström, A. & Jakobsson, M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 40, 4477–4482 (2013)
Acknowledgements
We thank P. Bellwood, J. Burger, P. Heggarty, M. Lipson, C. Renfrew, J. Diamond, S.Pääbo, R. Pinhasi and P. Skoglund for critical comments, and the Initiative for the Science of the Human Past at Harvard for organizing a workshop around the issues touched on by this paper. We thank S. Pääbo for support for establishing the ancient DNA facilities in Boston, and P. Skoglund for detecting the presence of two related individuals in our data set. We thank L. Orlando, T. S. Korneliussen, and C. Gamba for help in obtaining data. We thank Agilent Technologies and G. Frommer for help in developing the capture reagents. We thank C. Der Sarkissian, G. Valverde, L. Papac and B. Nickel for wet laboratory support. We thank archaeologists V. Dresely, R. Ganslmeier, O. Balanvosky, J. Ignacio Royo Guillén, A. Osztás, V. Majerik, T. Paluch, K. Somogyi and V.Voicsek for sharing samples and discussion about archaeological context. This research was supported by an Australian Research Council grant to W.H. and B.L. (DP130102158), and German Research Foundation grants to K.W.A. (Al 287/7-1 and 7-3, Al 287/10-1 and Al 287/14-1) and to H.M. (Me 3245/1-1 and 1-3). D.R. was supported by US National Science Foundation HOMINID grant BCS-1032255, US National Institutes of Health grant GM100233, and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
W.H., N.P., N.R., J.K., K.W.A. and D.R. supervised the study. W.H., E.B., C.E., M.F., S.F., R.G.P., F.H., V.K., A.K., M.K., P.K., H.M., O.M., V.M., N.N., S.L.P., R.R., M.A.R.G., C.R., A.S.-N., J.W., J.K., D.B., D.A., A.C., K.W.A. and D.R. assembled archaeological material, W.H., I.L., N.P., N.R., S.M., A.M. and D.R. analysed genetic data. I.L., N.P. and D.R. developed methods using f statistics for inferring admixture proportions. W.H., N.R., B.L., G.B., S.N., E.H., K.S. and A.M. performed wet laboratory ancient DNA work. I.L., N.R., S.M., B.L., Q.F., M.M. and D.R. developed the 390k capture reagent. W.H., I.L. and D.R. wrote the manuscript with help from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 2 Modelling Corded Ware as a mixture of N = 1, 2, or 3 ancestral populations.
a, The left column shows a histogram of raw f4 statistic residuals and on the right Z-scores for the best-fitting (lowest squared 2-norm of the residuals, or resnorm) model at each N. b, The data on the left show resnorm and on the right show the maximum |Z| score change for different N. c, resnorm of different N = 2 models. The set of outgroups used in this analysis in the terminology of Supplementary Information section 9 is ‘World Foci 15 + Ancients’.
Extended Data Figure 3 Modelling Europeans as mixtures of increasing complexity: N = 1 (EN), N = 2 (EN, WHG), N = 3 (EN, WHG, Yamnaya), N = 4 (EN, WHG, Yamnaya, Nganasan), N = 5 (EN, WHG, Yamnaya, Nganasan, BedouinB).
The residual norm of the fitted model (Supplementary Information section 9) and its changes are indicated.
Extended Data Figure 4 Geographic distribution of archaeological cultures and graphic illustration of proposed population movements / turnovers discussed in the main text.
a, Proposed routes of migration by early farmers into Europe ∼9,000−7000 years ago. b, Resurgence of hunter-gatherer ancestry during the Middle Neolithic 7,000−5,000 years ago. c, Arrival of steppe ancestry in central Europe during the Late Neolithic ∼4,500 years ago. White arrows indicate the two possible scenarios of the arrival of Indo-European language groups. Symbols of samples are identical to those in Fig. 1.
Supplementary information
Supplementary Information
This file contains Supplementary Information sections 1-11, see contents page for more details (PDF 31143 kb)
Supplementary Data
This file contains Supplementary Data 1. (XLSX 83 kb)
Supplementary Data
This file contains Supplementary Data 2a. (ZIP 12329 kb)
Supplementary Data
This file contains Supplementary Data 2b. (ZIP 14459 kb)
Supplementary Data
This file contains Supplementary Data 2c. (ZIP 9269 kb)
Supplementary Data
This file contains Supplementary Data 2d. (ZIP 2401 kb)
Rights and permissions
About this article
Cite this article
Haak, W., Lazaridis, I., Patterson, N. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). https://doi.org/10.1038/nature14317
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14317
This article is cited by
-
Differentiated adaptative genetic architecture and language-related demographical history in South China inferred from 619 genomes from 56 populations
BMC Biology (2024)
-
The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes
Scientific Data (2024)
-
Kinship practices at the early bronze age site of Leubingen in Central Germany
Scientific Reports (2024)
-
Human-mediated migration of plants, their pathogens and parasites
Journal of Plant Pathology (2024)
-
Population genomics of post-glacial western Eurasia
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.