Abstract
The aryl hydrocarbon receptor (AhR) is a highly conserved ligand-dependent transcription factor that senses environmental toxins and endogenous ligands, thereby inducing detoxifying enzymes and modulating immune cell differentiation and responses. We hypothesized that AhR evolved to sense not only environmental pollutants but also microbial insults. We characterized bacterial pigmented virulence factors, namely the phenazines from Pseudomonas aeruginosa and the naphthoquinone phthiocol from Mycobacterium tuberculosis, as ligands of AhR. Upon ligand binding, AhR activation leads to virulence factor degradation and regulated cytokine and chemokine production. The relevance of AhR to host defence is underlined by heightened susceptibility of AhR-deficient mice to both P. aeruginosa and M. tuberculosis. Thus, we demonstrate that AhR senses distinct bacterial virulence factors and controls antibacterial responses, supporting a previously unidentified role for AhR as an intracellular pattern recognition receptor, and identify bacterial pigments as a new class of pathogen-associated molecular patterns.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nguyen, L. P. & Bradfield, C. A. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 21, 102–116 (2008)
Schrenk, D. Impact of dioxin-type induction of drug-metabolizing enzymes on the metabolism of endo- and xenobiotics. Biochem. Pharmacol. 55, 1155–1162 (1998)
Guengerich, F. P. Cytochrome p450 and chemical toxicology. Chem. Res. Toxicol. 21, 70–83 (2008)
Casado, F. L., Singh, K. P. & Gasiewicz, T. A. The aryl hydrocarbon receptor: regulation of hematopoiesis and involvement in the progression of blood diseases. Blood Cells Mol. Dis. 44, 199–206 (2010)
Marlowe, J. L. & Puga, A. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell. Biochem. 96, 1174–1184 (2005)
Opitz, C. A. et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011)
Li, Y. et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640 (2011)
Lai, Z. W., Pineau, T. & Esser, C. Identification of dioxin-responsive elements (DREs) in the 5′ regions of putative dioxin-inducible genes. Chem. Biol. Interact. 100, 97–112 (1996)
Jensen, B. A., Leeman, R. J., Schlezinger, J. J. & Sherr, D. H. Aryl hydrocarbon receptor (AhR) agonists suppress interleukin-6 expression by bone marrow stromal cells: an immunotoxicology study. Environ. Health 2, 16 (2003)
Kimura, A. et al. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J. Exp. Med. 206, 2027–2035 (2009)
Quintana, F. J. et al. Control of Treg and Th17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008)
Veldhoen, M. et al. The aryl hydrocarbon receptor links Th17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008)
Apetoh, L. et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nature Immunol. 11, 854–861 (2010)
Gandhi, R. et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3+ regulatory T cells. Nature Immunol. 11, 846–853 (2010)
Kiss, E. A. et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565 (2011)
Qiu, J. et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104 (2012)
Motto, I. et al. New aryl hydrocarbon receptor homology model targeted to improve docking reliability. J. Chem. Inf. Model. 51, 2868–2881 (2011)
Rada, B. & Leto, T. L. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 21, 73–81 (2013)
Kerr, J. R. Phenazine pigments: antibiotics and virulence factors. Infect Dis. Rev. 29, 184–194 (2000)
Gardner, P. R. Superoxide production by the mycobacterial and pseudomonad quinoid pigments phthiocol and pyocyanine in human lung cells. Arch. Biochem. Biophys. 333, 267–274 (1996)
Lau, G. W., Hassett, D. J., Ran, H. & Kong, F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10, 599–606 (2004)
Wilson, R. et al. Pyocyanin and 1-hydroxyphenazine produced by Pseudomonas aeruginosa inhibit the beating of human respiratory cilia in vitro. J. Clin. Invest. 79, 221–229 (1987)
Denning, G. M. et al. Phenazine-1-carboxylic acid, a secondary metabolite of Pseudomonas aeruginosa, alters expression of immunomodulatory proteins by human airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 285, L584–L592 (2003)
Caldwell, C. C. et al. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am. J. Pathol. 175, 2473–2488 (2009)
Morrissey, J. P., Cullinane, M., Abbas, A., Mark, G. L. & O’Gara, F. Biosynthesis and regulation of anti-fungal metabolites by Pseudomonads, in Pseudomonas Vol III, (ed. by Ramos, J. L. ) 637–670 (Springer Verlag, 2004)
Hunter, R. C. et al. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am. J. Respir. Cell Mol. Biol. 47, 738–745 (2012)
Wilson, R. et al. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56, 2515–2517 (1988)
Wincent, E. et al. Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proc. Natl Acad. Sci. USA 109, 4479–4484 (2012)
Dietrich, L. E. et al. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61, 1308–1321 (2006)
Reszka, K. J. et al. Inactivation of the potent Pseudomonas aeruginosa cytotoxin pyocyanin by airway peroxidases and nitrite. Am. J. Physiol. Lung Cell. Mol. Physiol. 302, L1044–L1056 (2012)
Muller, M. Glutathione modulates the toxicity of, but is not a biologically relevant reductant for, the Pseudomonas aeruginosa redox toxin pyocyanin. Free Radic. Biol. Med. 50, 971–977 (2011)
O’Malley, Y. Q. et al. Pseudomonas aeruginosa pyocyanin directly oxidizes glutathione and decreases its levels in airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 287, L94–L103 (2004)
Nebert, D. W., Puga, A. & Vasiliou, V. Role of the Ah receptor and the dioxin-inducible [Ah] gene battery in toxicity, cancer, and signal transduction. Ann. NY Acad. Sci. 685, 624–640 (1993)
Lavoie, E. G., Wangdi, T. & Kazmierczak, B. I. Innate immune responses to Pseudomonas aeruginosa infection. Microbes Infect. 13, 1133–1145 (2011)
Goldberg, J. B. & Pier, G. B. The role of the CFTR in susceptibility to Pseudomonas aeruginosa infections in cystic fibrosis. Trends Microbiol. 8, 514–520 (2000)
Schuster, M. G. & Norris, A. H. Community-acquired Pseudomonas aeruginosa pneumonia in patients with HIV infection. AIDS 8, 1437–1441 (1994)
Kobayashi, Y. The role of chemokines in neutrophil biology. Front. Biosci. 13, 2400–2407 (2008)
Cheon, H. et al. Signaling pathway for 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced TNF-α production in differentiated THP-1 human macrophages. Exp. Mol. Med. 39, 524–534 (2007)
Vogel, C. F. & Matsumura, F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-κB family. Biochem. Pharmacol. 77, 734–745 (2009)
Eisele, N. A. & Anderson, D. M. Host defense and the airway epithelium: frontline responses that protect against bacterial invasion and pneumonia. J. Pathogens http://dx.doi.org/10.4061/2011/249802 (2011)
Lee, Y. C. et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced MUC5AC expression: aryl hydrocarbon receptor-independent/EGFR/ERK/p38-dependent SP1-based transcription. Am. J. Respir. Cell Mol. Biol. 45, 270–276 (2011)
Dorhoi, A., Reece, S. T. & Kaufmann, S. H. For better or for worse: the immune response against Mycobacterium tuberculosis balances pathology and protection. Immunol. Rev. 240, 235–251 (2011)
Weiner, J., III et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS ONE 7, e40221 (2012)
Mavrodi, D. V., Blankenfeldt, W. & Thomashow, L. S. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44, 417–445 (2006)
Pierson, L. S., III & Pierson, E. A. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 86, 1659–1670 (2010)
Poltorak, A. et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 (1998)
Medzhitov, R., Preston-Hurlburt, P. & Janeway, C. A., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 (1997)
Echenique, P. & Alonso, J. L. A mathematical and computational review of Hartree–Fock SCF methods in quantum chemistry. Mol. Phys. 105, 3057–3098 (2010)
Schmidt, M. W. et al. General atomic and molecular electronic structure system. J. Comput.. Chem. 14, 1347–1363 (1993)
Barouki, R., Aggerbeck, M., Aggerbeck, L. & Coumoul, X. The aryl hydrocarbon receptor system. Drug Metabol. Drug Interact. 27, 3–8 (2012)
Smith, K. J. et al. Identification of a high-affinity ligand that exhibits complete aryl hydrocarbon receptor antagonism. J. Pharmacol. Exp. Ther. 338, 318–327 (2011)
Bisson, W. H. et al. Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J. Med. Chem. 52, 5635–5641 (2009)
Pandini, A. et al. Structural and functional characterization of the aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis. Biochemistry 46, 696–708 (2007)
Fraccalvieri, D. et al. Comparative analysis of homology models of the AH receptor ligand binding domain: verification of structure-function predictions by site-directed mutagenesis of a nonfunctional receptor. Biochemistry 52, 714–725 (2013)
Farmahin, R. et al. Amino acid sequence of the ligand-binding domain of the aryl hydrocarbon receptor 1 predicts sensitivity of wild birds to effects of dioxin-like compounds. Toxicol. Sci. 131, 139–152 (2013)
Kaiser, C. M. et al. Real-time observation of trigger factor function on translating ribosomes. Nature 444, 455–460 (2006)
Lakowicz, J. R. Principles of Fluorescence Spectroscopy 3rd edn (Springer, 2006)
Fan, M. Q. et al. Recombinant expression of aryl hydrocarbon receptor for quantitative ligand-binding analysis. Anal. Biochem. 384, 279–287 (2009)
Cheng, Y. & Prusoff, W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973)
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1–0034.11 (2002)
Klockgether, J. et al. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 192, 1113–1121 (2010)
Clausen, B. E. et al. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 (1999)
Munder, A. et al. Acute intratracheal Pseudomonas aeruginosa infection in cystic fibrosis mice is age-independent. Respir. Res. 12, 148 (2011)
Kursar, M. et al. Cutting edge: regulatory T cells prevent efficient clearance of Mycobacterium tuberculosis. J. Immunol. 178, 2661–2665 (2007)
Churchill, G. A. Fundamentals of experimental design for cDNA microarrays. Nature Genet. 32 (Suppl). 490–495 (2002)
Nebert, D. W. et al. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 59, 65–85 (2000)
Tijet, N. et al. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 69, 140–153 (2006)
Acknowledgements
The authors are highly grateful to J. Welch for his contribution in the early stage of research, and for support in AhR ligand screening, A. Zychlinsky and L. E. Dietrich for discussions, H.-G. Hoymann for support in lung function measurements and interpretation of results, U. Klemm for mouse breeding and M. L. Grossman for excellent support in preparing the manuscript. Ahr–/–mice were provided by B. Stockinger and shRNA constructs by D. Krastev and F. Buchholz. The PA14 WT2 and PA14 Δphz1/2 were a gift from D. K. Newman and L. E. Dietrich. M. Kolbe acknowledges grant support from the European Union’s Seventh Framework Programmes (EU-FP7/2007-2013). S.H.E.K. acknowledges grant support from the European Union’s Seventh Framework Programmes (EU-FP7) NEWTBVAC (Health-F3-2009-241745) and PHAGOSYS (Health-F4-2008-223451).
Author information
Authors and Affiliations
Contributions
P.M.-A., E.H., K.F., A. Dorhoi and S.H.E.K. conceived and designed the study and wrote the manuscript. P.M.-A., E.H., K.F. and A. Dorhoi designed and performed experiments and data analysis. P.C. and M.D. performed Mycobacterium lipid fractionation. A.M. and B.T. designed and performed Pseudomonas spirometry studies. U.G.-B., M. Klemm, A.-B.K. and S.B. provided technical help for in vitro and in vivo experiments. R.H. performed and analysed HPLC experiments. A.K., G.K. and H.O. performed virtual docking studies. S.F. discussed experiments. T.S., V.B. and C.G. performed and discussed experiments. H.-J.M. performed and analysed microarray experiments. M. Kolbe, N.B., J.F., A. Diehl and H.O. performed binding studies. All authors commented on the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 AhR binding, activation and cell viability.
a, Chemical structures of phenazine-1-carboxylic acid (PCA) and phenazine-1-carboxamide (PCN). b, In silico docking of PCA and PCN into the ligand-binding pocket (yellow surface) of AhR. Hydrophilic residues (magenta), aromatic and hydrophobic residues (green). c, Intrinsic tryptophan fluorescence quenching of purified AhR1–417 or mock transfected control titrated with increasing concentrations of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) or 1-hydroxyphenazine (1-HP). d–f, Luciferase activity of AhR reporter cells after 24 h stimulation. d, THP-1 cells stimulated with 50 μM of PCA or PCN. e, H358 cells stimulated with TCDD (10 nM), pyocyanin (Pyo, 50 μM), 1-HP (50 μM) or phthiocol (Pht, 50 µM). f, AhR activation of THP-1 AhR reporter cells upon stimulation with different concentrations of known AhR ligands (TCDD, 6-formylindolo[3,2-b] carbazole (FICZ), β-naphthoflavone or kynurenic acid (Kyn) or bacterial pigments (Pyo, 1-HP or Pht). g, Cell viability assessed after 24 h stimulation with 50 μM of Pyo, 1-HP and Pht. h, Luciferase activity of THP-1 AhR reporter cells stimulated for 24 h with 50 μM of Pyo, 1-HP, Pht and lipopolysaccharide (LPS, 1 μg ml−1), flagellin (100 ng ml−1) or CpG oligodeoxynucleotides (ODN2006, 5 μM). c, Representative of at least two experiments; d–h, cumulative data of at least three experiments, mean + s.e.m.; d–h, Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.001.
Extended Data Figure 2 Bacterial pigments induce transcription of canonical AhR pathway genes.
a, qRT–PCR of CYP1A1, AHRR and CYP1B1 in different cells after stimulation with 50 μM of Pyo, 1-HP and Pht. b, AhR gene knockdown (KD) efficiency following infection with lentivirus encoding a pool of AhR-specific shRNAs. Cells transduced with a non-targeting Scramble shRNA were considered as reference control. c, qRT–PCR of CYP1A1 in H358 Scramble and AhR-KD cells after 24 h stimulation with 50 μM of Pyo, 1-HP and Pht. a–c, Cumulative data of at least three experiments, mean + s.e.m.; a–c, Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Extended Data Figure 3 Bacterial pigments induce global AhR signalling in pneumocytes.
Microarray analysis of A549 Scramble cells upon stimulation with virulence factors. Cells were treated with 50 μM of the different bacterial pigmented virulence factors or DMSO as control for 24 h. RNA was collected and subjected to microarray analysis. a, List of genes differentially expressed (P < 0.00001) upon stimulation of cells with the different ligands, as compared to DMSO. b, Top 20 canonical pathways predicted by Ingenuity pathway analysis software to be differentially regulated upon stimulation. Up, upregulated. Down, downregulated. Blue bars (left, y axis) depict –log P values calculated by Fisher’s exact test whereas yellow line (right, y axis) represents the ratio between the number of genes in a given pathway compared to the total number of genes in that pathway.
Extended Data Figure 4 AhR activation in THP-1 cells in the absence of Trp or CYP1A1.
a, Luciferase activity of AhR reporter in THP-1 cells stimulated with TCDD (10 nM), FICZ (20 nM), Pyo (50 μM), 1-HP (50 μM) or Pht (50 µM) for 24 h in the absence of Trp. b, CYP1A1 gene KD efficiency in THP-1 cells. Cells were transduced using lentivirus encoding a pool of CYP1A1-specific shRNAs. Cells transduced with a non-targeting Scramble shRNA were considered as reference control. c, d, qRT–PCR of AHRR in THP-1 in Scramble control (c) and CYP1A1-KD (d) cells after 24 h stimulation with 10 nM TCDD or 50 μM of Pyo, 1-HP and Pht. a–d, Cumulative data of at least three experiments, mean + s.e.m.; a–d, Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Extended Data Figure 5 P. aeruginosa PA14 bacterial growth, phenazine concentrations and AhR activation.
Different PA14 mutants (09480 and Δphz1/2) and parental WT controls (WT1 and WT2, respectively) were tested. a, Bacterial density and Pyo concentration in the filtered supernatants were determined at different time points. Means of bacterial density and Pyo concentration are shown. Full lines represent bacterial growth, and dashed lines depict Pyo concentration for each strain. b, HPLC analysis of different phenazines (Pyo, 1-HP, PCA and PCN) present in the supernatants of P. aeruginosa PA14 WT1 and mutant PA14 09480 strains. c, d, Filtered supernatants from PA14 mutants and parental WT controls were used to stimulate THP-1 AhR reporter cell line for 24 h. Luciferase activity was measured and normalized to non-stimulated cells (control). a, Representative of at least three experiments; b, representative of at least three experiments, mean + s.d.; c, d, cumulative data of three experiments, mean + s.e.m. c, d, Student’s t-test. **P < 0.01.
Extended Data Figure 6 Degradation of bacterial pigments upon AhR activation.
a, b, HPLC of Pyo (a) and Pht (b) degradation in supernatants of A549 Scramble and AhR-KD cells 48 h after stimulation. a, Arrows depict new peaks emerging at 368 nm at 1.21, 1.31 and 1.42 min with phenazine-like characteristics, suggesting formation of a new metabolite(s) from Pyo. Higher levels of these metabolite(s) are observed in supernatants of scramble cells challenged with Pyo, suggesting an AhR dependent role. These peaks were not detected in supernatants of unstimulated cells. b, Decreased levels of native Pht (arrow) are detected in supernatants of A549 Scramble cells, 48 h after challenge with Pht. c, Expression of the AhR gene battery: Phase I and Phase II xenobiotic metabolizing enzymes (XME)2,33,66,67. A549 cells (Scramble and AhR-KD) were stimulated for 24 h with different bacterial ligands or DMSO as control. RNA was extracted and microarray analysis was performed. The table depicts fold induction of different XME upon stimulation, as compared to DMSO control in different cell lines tested. The remaining differently regulated genes in A549 cells (Scramble and AhR-KD) upon stimulation are depicted in the Supplementary Tables 4 and 5. n.d., not detected. a, b, Representative of three experiments.
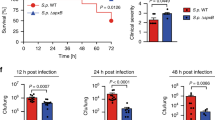
Extended Data Figure 7 Ahr–/– mice are more susceptible to P. aeruginosa infection.
WT and AhR-deficient (Ahr–/–) mice were infected intratracheally (i.t.) with 107 c.f.u. (a), 2 × 105 c.f.u. (b) or 4 × 106 c.f.u. (c) of P. aeruginosa PAO1. a, Body temperature. b, Lung function evaluated by non-invasive head-out spirometry. Spirometric curves depict time course of tidal volume (total volume inspired and expired in one breath), expiratory flow at 50% expiration (EF50) and rate (breaths per min). c, Bacterial loads. a, Cumulative data of two experiments, mean ± s.e.m.; b, representative of at least two experiments, mean ± s.d.; c, representative of at least two experiments, median; a, b two-way ANOVA; c, Mann–Whitney U- test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Extended Data Figure 8 Cellular, cytokine and microarray analysis of P. aeruginosa-infected mice.
a, b, d–h, WT and Ahr–/– mice were infected i.t. with 4 × 106 c.f.u. of PAO1. a, b, Frequencies and absolute numbers of myeloid cells from lung and bronchoalveolar lavage fluid (BALF). c, Frequencies and absolute numbers of myeloid cells from lungs of naive animals. d, e, Frequencies of lymphoid cells in lungs from naive mice. f, Cytokine and chemokine abundance in BALF from P. aeruginosa-infected mice. g, h, Microarray analysis of BALF samples from infected WT mice, as compared to PBS control. g, List of genes differentially expressed (P < 0.00001) upon infection of mice with P. aeruginosa PAO1. h, Top 20 canonical pathways predicted by Ingenuity pathway analysis software to be differentially regulated upon infection. Up, upregulated; down, downregulated. Blue bars (left, y axis) depict –log P values calculated by Fisher's exact test. Yellow line (right, y axis) represents the ratio between number of genes in a given pathway compared to total number of genes in that pathway. a, b, d, e, Representative of two experiments, mean + s.d.; c, cumulative data of two experiments, mean + s.e.m.; f, cumulative data of two experiments, median + interquartile range; a–d, two-way ANOVA; e, f, (Mann–Whitney U- test). *P < 0.05; **P < 0.01; ****P < 0.0001.
Extended Data Figure 9 M. tuberculosis infection of mice and human primary macrophages.
a–d, WT and Ahr–/–mice were aerosol-infected with M. tuberculosis H37Rv (low-dose, 100 c.f.u.). a, b, Tissue damage. a, Lactate dehydrogenase (LDH) activity in serum. b, Hematoxylin and eosin staining of lung. Scale bars, 500 μm. c, Flow cytometry analysis of lymphoid cells. d, Cytokine and chemokine abundances in lung homogenates. e, AhR activation by M. tuberculosis in human primary macrophages. qRT–PCR of CYP1A1 and AHRR after infection with H37Rv (m.o.i. = 5). a, c Representative of two experiments, mean + s.d.; b, representative of two experiments; d, cumulative data of two experiments, median + interquartile ranges; e, n = 3 donors, mean + s.d.; a, c, two-way ANOVA; d, Mann–Whitney U-test; e, Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Figure 10 AhR activation by M. tuberculosis lipid fractions.
a, Luciferase activity of AhR reporter in THP-1 cells stimulated with different M. tuberculosis preparations (cell wall, membrane and cytosolic fractions, total lipids, mannosilated lipoarabinomannan (ManLAM), trehelose dimycolate (TDM), antigen 85 (Ag85), Ag85B, early secretory antigenic target 6 (ESAT6) and 10 kDa culture filtrate antigen (CFP10)). b, Flow chart of the sequence of M. tuberculosis lipid fractionation by chromatography on a Florisil column and identification of AhR-activating fractions. c, Luciferase activity of AhR reporter in THP-1 cells stimulated for 24 h with different M. tuberculosis lipid fractions. d, Gas chromatography coupled mass spectrometry (GC/MS) identification of Pht in AhR-activating versus -nonactivating lipid fractions. In the active fraction, an elution peak was observed at the same retention time as Pht, showing the same mass fragmentation pattern (m/z 188, 160, 131, 105, 77).
Supplementary information
Supplementary Information
This file contains Supplementary Text. (PDF 135 kb)
Supplementary Data
This zipped file contains Supplementary Tables 1-11. (ZIP 1290 kb)
Rights and permissions
About this article
Cite this article
Moura-Alves, P., Faé, K., Houthuys, E. et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature 512, 387–392 (2014). https://doi.org/10.1038/nature13684
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13684
This article is cited by
-
Endothelial AHR activity prevents lung barrier disruption in viral infection
Nature (2023)
-
The aryl hydrocarbon receptor regulates expression of mucosal trafficking receptor GPR15
Mucosal Immunology (2021)
-
The role of DNA-binding and ARNT dimerization on the nucleo-cytoplasmic translocation of the aryl hydrocarbon receptor
Scientific Reports (2021)
-
Aryl hydrocarbon receptor (AHR) functions in infectious and sterile inflammation and NAD+-dependent metabolic adaptation
Archives of Toxicology (2021)
-
Pulmonary paracoccidioidomycosis in AhR deficient hosts is severe and associated with defective Treg and Th22 responses
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.