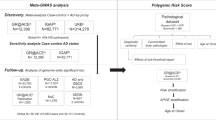
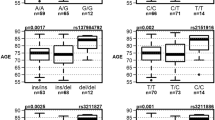
Abstract
The prevalence of dementia in the Western world in people over the age of 60 has been estimated to be greater than 5%, about two-thirds of which are due to Alzheimer’s disease1,2,3,4. The age-specific prevalence of Alzheimer’s disease nearly doubles every 5 years after age 65, leading to a prevalence of greater than 25% in those over the age of 90 (ref. 3). Here, to search for low-frequency variants in the amyloid-β precursor protein (APP) gene with a significant effect on the risk of Alzheimer’s disease, we studied coding variants in APP in a set of whole-genome sequence data from 1,795 Icelanders. We found a coding mutation (A673T) in the APP gene that protects against Alzheimer’s disease and cognitive decline in the elderly without Alzheimer’s disease. This substitution is adjacent to the aspartyl protease β-site in APP, and results in an approximately 40% reduction in the formation of amyloidogenic peptides in vitro. The strong protective effect of the A673T substitution against Alzheimer’s disease provides proof of principle for the hypothesis that reducing the β-cleavage of APP may protect against the disease. Furthermore, as the A673T allele also protects against cognitive decline in the elderly without Alzheimer’s disease, the two may be mediated through the same or similar mechanisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
01 August 2012
A minor error relating to the rs63750847 variant in ref. 15 was corrected.
References
Ferri, C. P. et al. Global prevalence of dementia: a Delphi consensus study. Lancet 366, 2112–2117 (2005)
Plassman, B. L. et al. Prevalence of dementia in the United States: the aging, demographics, and memory study. Neuroepidemiology 29, 125–132 (2007)
Qiu, C., Kivipelto, M. & von Strauss, E. Epidemiology of Alzheimer’s disease: occurrence, determinants, and strategies toward intervention. Dialogues Clin. Neurosci. 11, 111–128 (2009)
Reitz, C., Brayne, C. & Mayeux, R. Epidemiology of Alzheimer disease. Nature reviews . Neurology 7, 137–152 (2011)
Glenner, G. G. & Wong, C. W. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 (1984)
Masters, C. L. et al. Neuronal origin of a cerebral amyloid: neurofibrillary tangles of Alzheimer’s disease contain the same protein as the amyloid of plaque cores and blood vessels. EMBO J. 4, 2757–2763 (1985)
Zhang, Y. W., Thompson, R., Zhang, H. & Xu, H. APP processing in Alzheimer’s disease. Mol. Brain 4, 3 (2011)
Hussain, I. et al. Identification of a novel aspartic protease (Asp 2) as β-secretase. Mol. Cell. Neurosci. 14, 419–427 (1999)
Sinha, S. et al. Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature 402, 537–540 (1999)
Vassar, R. et al. β-Secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 (1999)
Yan, R. et al. Membrane-anchored aspartyl protease with Alzheimer’s disease β-secretase activity. Nature 402, 533–537 (1999)
Hardy, J. C. & Crook, R. APP mutations table. http://www.alzforum.org/res/com/mut/app/table1.asp (2010)
Cruts, M. Alzheimer’s disease and frontotemporal dementia mutation database. http://www.molgen.ua.ac.be/ADMutations (2012)
St George-Hyslop, P. H. Molecular genetics of Alzheimer’s disease. Biol. Psychiatry 47, 183–199 (2000)
Cruchaga, C. et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PLoS ONE 7, e31039 (2012)
Kong, A. et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nature Genet. 40, 1068–1075 (2008)
Kong, A. et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467, 1099–1103 (2010)
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 39, 906–913 (2007)
Sulem, P. et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nature Genet. 43, 1127–1130 (2011)
Peacock, M. L., Warren, J. T., Jr, Roses, A. D. & Fink, J. K. Novel polymorphism in the A4 region of the amyloid precursor protein gene in a patient without Alzheimer’s disease. Neurology 43, 1254–1256 (1993)
Exome Variant Server. NHLBI Exome Sequencing Project (ESP). http://evs.gs.washington.edu/EVS/ (2012)
Di Fede, G. et al. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science 323, 1473–1477 (2009)
Giaccone, G. et al. Neuropathology of the recessive A673V APP mutation: Alzheimer disease with distinctive features. Acta Neuropathol. 120, 803–812 (2010)
Citron, M. et al. Mutation of the β-amyloid precursor protein in familial Alzheimer’s disease increases β-protein production. Nature 360, 672–674 (1992)
Gruninger-Leitch, F., Schlatter, D., Kung, E., Nelbock, P. & Dobeli, H. Substrate and inhibitor profile of BACE (β-secretase) and comparison with other mammalian aspartic proteases. J. Biol. Chem. 277, 4687–4693 (2002)
Tomasselli, A. G. et al. Employing a superior BACE1 cleavage sequence to probe cellular APP processing. J. Neurochem. 84, 1006–1017 (2003)
Morris, J. N. et al. MDS Cognitive Performance Scale. J. Gerontol. 49, M174–M182 (1994)
Holm, H. et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nature Genet. 43, 316–320 (2011)
Acknowledgements
We would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Author information
Authors and Affiliations
Contributions
The study was designed and results were interpreted by T.J., J.K.A., H.S., R.J.W. and K.S. Sequence data analysis was carried out by T.J., S.S., P.S., A.K., T.B., R.R.G., T.W.B. and D.G. Subject recruitment, phenotype analysis and biological material collection was organized and carried out by J.S., P.V.J., S.B., G.B., O.A.A., E.G.J. and A.P. Sequencing and genotyping was supervised by J.H., O.T.M. and U.T. Cell line experiments and BACE1 cleavage assays were carried out and analysed by J.K.A., J.M., K.H., Y. Lu, Y. Liu, A.G. and R.J.W. The paper was drafted by T.J., J.K.A., R.J.W. and K.S. All authors contributed to the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
Authors from deCODE and Genentech are employees of deCODE genetics, ehf and Genentech, respectively.
Supplementary information
Supplementary Information
This file contains Supplementary Text and Data, Supplementary Tables 1-2, Supplementary Figures 1-4 and Supplementary References. (PDF 998 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Jonsson, T., Atwal, J., Steinberg, S. et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488, 96–99 (2012). https://doi.org/10.1038/nature11283
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11283
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.