Abstract
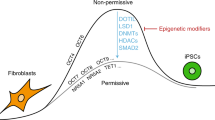
Generation of induced pluripotent stem cells (iPSCs) by somatic cell reprogramming involves global epigenetic remodelling1. Whereas several proteins are known to regulate chromatin marks associated with the distinct epigenetic states of cells before and after reprogramming2,3, the role of specific chromatin-modifying enzymes in reprogramming remains to be determined. To address how chromatin-modifying proteins influence reprogramming, we used short hairpin RNAs (shRNAs) to target genes in DNA and histone methylation pathways, and identified positive and negative modulators of iPSC generation. Whereas inhibition of the core components of the polycomb repressive complex 1 and 2, including the histone 3 lysine 27 methyltransferase EZH2, reduced reprogramming efficiency, suppression of SUV39H1, YY1 and DOT1L enhanced reprogramming. Specifically, inhibition of the H3K79 histone methyltransferase DOT1L by shRNA or a small molecule accelerated reprogramming, significantly increased the yield of iPSC colonies, and substituted for KLF4 and c-Myc (also known as MYC). Inhibition of DOT1L early in the reprogramming process is associated with a marked increase in two alternative factors, NANOG and LIN28, which play essential functional roles in the enhancement of reprogramming. Genome-wide analysis of H3K79me2 distribution revealed that fibroblast-specific genes associated with the epithelial to mesenchymal transition lose H3K79me2 in the initial phases of reprogramming. DOT1L inhibition facilitates the loss of this mark from genes that are fated to be repressed in the pluripotent state. These findings implicate specific chromatin-modifying enzymes as barriers to or facilitators of reprogramming, and demonstrate how modulation of chromatin-modifying enzymes can be exploited to more efficiently generate iPSCs with fewer exogenous transcription factors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Change history
29 March 2012
Author name for B.O.M. was corrected.
References
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006)
Hawkins, R. D. et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6, 479–491 (2010)
Mikkelsen, T. S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007)
Park, I.-H. et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 (2008)
Margueron, R. & Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011)
Pereira, C. F. et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6, 547–556 (2010)
Shi, Y. et al. Transcriptional repression by YY1, a human GLI-Krüippel-related protein, and relief of repression by adenovirus E1A protein. Cell 67, 377–388 (1991)
Schotta, G., Ebert, A. & Reuter, G. S. U. (VAR)3–9 is a conserved key function in heterochromatic gene silencing. Genetica 117, 149–158 (2003)
Jones, B. et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 4, e1000190 (2008)
Okada, Y. et al. hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178 (2005)
Daigle, S. R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011)
Bernt, K. M. et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78 (2011)
Carey, B. W. et al. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nature Methods 7, 56–59 (2010)
Boyer, L. A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005)
Mikkelsen, T. S. et al. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 (2008)
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007)
Charafe-Jauffret, E. et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 25, 2273–2284 (2006)
Onder, T. T. et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 68, 3645–3654 (2008)
Taube, J. H. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl Acad. Sci. USA 107, 15449–15454 (2010)
Samavarchi-Tehrani, P. et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 (2010)
Stulemeijer, I. J. e. t. a. l. Dot1 binding induces chromatin rearrangements by histone methylation-dependent and -independent mechanisms. Epigenetics chromatin 4, 2 (2011)
Olson, A. et al. RNAi Codex: a portal/database for short-hairpin RNA (shRNA) gene-silencing constructs. Nucleic Acids Res. 34, D153–D157 (2006)
Schlabach, M. R. et al. Cancer proliferation gene discovery through functional genomics. Science 319, 620–624 (2008)
Zaehres, H. et al. High-efficiency RNA interference in human embryonic stem cells. Stem Cells 23, 299–305 (2005)
Park, I.-H. et al. Generation of human-induced pluripotent stem cells. Nature Protocols 3, 1180–1186 (2008)
Yu, J. et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 (2009)
Chan, E. M. et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nature Biotechnol. 27, 1033–1037 (2009)
Loewer, S. et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature Genet. 42, 1113–1117 (2010)
Langmead, B. et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Subramanian, A. et al. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23, 3251–3253 (2007)
Acknowledgements
We thank G. Hu and S. J. Elledge for providing the MSCV-PM vector, K. Ng and M. W. Lensch for teratoma injections and assessment and S. Loewer for discussions. We also thank E. Olhava and Epizyme Inc. for synthesizing and providing the DOT1L inhibitor, EPZ004777. G.Q.D. is an investigator of the Howard Hughes Medical Institute. Research was funded by grants from the US National Institutes of Health (NIH) to S.A.A. (CA140575) and G.Q.D., and the CHB Stem Cell Program.
Author information
Authors and Affiliations
Contributions
T.T.O. performed project planning, experimental work, data interpretation and preparation of the manuscript. N.K., A.C, N.Z., J.U. and B.O.M. performed experimental work. P.C. and A.U.S. participated in data analysis. K.M.B. and S.A.A. provided critical materials and participated in the preparation of the manuscript. P.B.G. and E.S.L., participated in data acquisition, data interpretation and preparation of the manuscript. G.Q.D. supervised research and participated in project planning, data interpretation and preparation of the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.A.A. is a consultant for Epizyme Inc. G.Q.D. is a member of the scientific advisory boards and holds stock in or receives consulting fees from the following companies: Johnson & Johnson, Verastem, Epizyme, iPierian, Solasia KK and MPM Capital, LLP.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-22. (PDF 1698 kb)
Supplementary Table 1
This table contains a list of all the shRNA sequences used. (XLS 21 kb)
Supplementary Table 2
This table contains a list of qRT-PCR primers used. (XLS 11 kb)
Supplementary Table 3
This table contains genes upregulated and downregulated upon Dot1L inhibition during reprogramming based on gene expression profiling. (XLS 56 kb)
Supplementary Table 4
This table contains the enrichment scores for H3K79me2 and H3K27me3 Chip-seq and lists of genes significantly enriched in the indicated cell populations. (XLS 6867 kb)
Supplementary Table 5
This table shows gene sets significantly enriched in the gene set overlap analysis. (XLS 635 kb)
Rights and permissions
About this article
Cite this article
Onder, T., Kara, N., Cherry, A. et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598–602 (2012). https://doi.org/10.1038/nature10953
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10953
This article is cited by
-
Canalizing cell fate by transcriptional repression
Molecular Systems Biology (2024)
-
B1 SINE-binding ZFP266 impedes mouse iPSC generation through suppression of chromatin opening mediated by reprogramming factors
Nature Communications (2023)
-
H3K36 methylation maintains cell identity by regulating opposing lineage programmes
Nature Cell Biology (2023)
-
A fast chemical reprogramming system promotes cell identity transition through a diapause-like state
Nature Cell Biology (2023)
-
The NuRD complex cooperates with SALL4 to orchestrate reprogramming
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.