Abstract
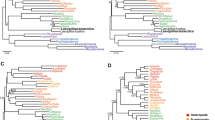
Molluscs (snails, octopuses, clams and their relatives) have a great disparity of body plans and, among the animals, only arthropods surpass them in species number. This diversity has made Mollusca one of the best-studied groups of animals, yet their evolutionary relationships remain poorly resolved1. Open questions have important implications for the origin of Mollusca and for morphological evolution within the group. These questions include whether the shell-less, vermiform aplacophoran molluscs diverged before the origin of the shelled molluscs (Conchifera)2,3,4 or lost their shells secondarily. Monoplacophorans were not included in molecular studies until recently5,6, when it was proposed that they constitute a clade named Serialia together with Polyplacophora (chitons), reflecting the serial repetition of body organs in both groups5. Attempts to understand the early evolution of molluscs become even more complex when considering the large diversity of Cambrian fossils. These can have multiple dorsal shell plates and sclerites7,8,9,10 or can be shell-less but with a typical molluscan radula and serially repeated gills11. To better resolve the relationships among molluscs, we generated transcriptome data for 15 species that, in combination with existing data, represent for the first time all major molluscan groups. We analysed multiple data sets containing up to 216,402 sites and 1,185 gene regions using multiple models and methods. Our results support the clade Aculifera, containing the three molluscan groups with spicules but without true shells, and they support the monophyly of Conchifera. Monoplacophora is not the sister group to other Conchifera but to Cephalopoda. Strong support is found for a clade that comprises Scaphopoda (tusk shells), Gastropoda and Bivalvia, with most analyses placing Scaphopoda and Gastropoda as sister groups. This well-resolved tree will constitute a framework for further studies of mollusc evolution, development and anatomy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Primary accessions
Sequence Read Archive
Data deposits
Illumina and 454 reads have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under accession number SRA044948. Sanger reads for Laevipilina hyalina have been deposited in the NCBI Trace Archive under the sequencing centre name BUDL with TI range 2317135955-2317139410. The assembled data, matrices and trees have been deposited in Dryad (http://dx.doi.org/10.5061/dryad.24cb8).
Change history
28 November 2012
Nature 480, 364–367 (2011); doi:10.1038/nature10526 In this Letter, we investigated the evolutionary relationships of molluscs with multigene data sets assembled from new transcriptome data and published genomes and transcriptomes. Since publishing these results, examination of our gene sequence matrix by others revealed that all instances of six amino acids (E, F, I, L, P and Q) were replaced by ambiguous characters in our super matrix.
References
Ponder, W. F., Lindberg, D. R., eds. Phylogeny and Evolution of the Mollusca (Univ. California Press, 2008)
Salvini-Plawen, L. V. & Steiner, G. in Origin and Evolutionary Radiation of the Mollusca (ed. Taylor, J. D. ) 29–51 (Oxford Univ. Press, 1996)
Scheltema, A. H. in Origin and Evolutionary Radiation of the Mollusca (ed. Taylor, J. D. ) 53–58 (Oxford Univ. Press, 1996)
Haszprunar, G. Is the Aplacophora monophyletic? A cladistic point of view. Am. Malacol. Bull. 15, 115–130 (2000)
Giribet, G. et al. Evidence for a clade composed of molluscs with serially repeated structures: monoplacophorans are related to chitons. Proc. Natl Acad. Sci. USA 103, 7723–7728 (2006)
Wilson, N. G., Rouse, G. W. & Giribet, G. Assessing the molluscan hypothesis Serialia (Monoplacophora + Polyplacophora) using novel molecular data. Mol. Phylogenet. Evol. 54, 187–193 (2010)
Vinther, J. & Nielsen, C. The Early Cambrian Halkieria is a mollusc. Zool. Scr. 34, 81–89 (2005)
Scheltema, A. H., Kerth, K. & Kuzirian, A. M. Original molluscan radula: comparisons among Aplacophora, Polyplacophora, Gastropoda, and the Cambrian fossil Wiwaxia corrugata. J. Morphol. 257, 219–245 (2003)
Morris, S. C. & Caron, J. B. Halwaxiids and the early evolution of the lophotrochozoans. Science 315, 1255–1258 (2007)
Sutton, M. D., Briggs, D. E. G., Siveter, D. J. & Siveter, D. J. Computer reconstruction and analysis of the vermiform mollusc Acaenoplax hayae from the Herefordshire Lagerstätte (Silurian, England), and implications for molluscan phylogeny. Palaeontology 47, 293–318 (2004)
Caron, J.-B., Scheltema, A., Schander, C. & Rudkin, D. A soft-bodied mollusc with radula from the Middle Cambrian Burgess Shale. Nature 442, 159–163 (2006)
Passamaneck, Y. J., Schander, C. & Halanych, K. M. Investigation of molluscan phylogeny using large-subunit and small-subunit nuclear rRNA sequences. Mol. Phylogenet. Evol. 32, 25–38 (2004)
Aktipis, S. W., Giribet, G., Lindberg, D. R. & Ponder, W. F. in Phylogeny and Evolution of the Mollusca (eds Ponder, W. F. & Lindberg, D. R. ) 201–237 (Univ. California Press, 2008)
Giribet, G. & Distel, D. L. in Molecular Systematics and Phylogeography of Mollusks (eds Lydeard, C. & Lindberg, D. R. ) 45–90 (Smithsonian Books, 2003)
Rokas, A., Krüger, D. & Carroll, S. B. Animal evolution and the molecular signature of radiations compressed in time. Science 310, 1933–1938 (2005)
Hejnol, A. et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. B 276, 4261–4270 (2009)
Dunn, C. W. et al. Broad taxon sampling improves resolution of the animal tree of life. Nature 452, 745–749 (2008)
Prendini, L. Species or supraspecific taxa as terminals in cladistic analysis? Groundplans versus exemplars revisited. Syst. Biol. 50, 290–300 (2001)
Vendrasco, M. J., Wood, T. E. & Runnegar, B. N. Articulated Palaeozoic fossil with 17 plates greatly expands disparity of early chitons. Nature 429, 288–291 (2004)
Nielsen, C., Haszprunar, G., Ruthensteiner, B. & Wanninger, A. Early development of the aplacophoran mollusc Chaetoderma. Acta Zool. 88, 231–247 (2007)
Wanninger, A., Koop, D., Moshel-Lynch, S. & Degnan, B. M. in Phylogeny and Evolution of the Mollusca (eds Ponder, W. F. & Lindberg, D. R.) 427–445 (Univ. California Press, 2008)
Yochelson, E. L. An alternative approach to the interpretation of the phylogeny of ancient mollusks. Malacologia 17, 165–191 (1978)
Runnegar, B. in Origin and Evolutionary Radiation of the Mollusca (ed. Taylor, J. D. ) 77–87 (Oxford Univ. Press, 1996)
Yochelson, E. L., Flower, R. H. & Webers, G. F. The bearing of the new Late Cambrian monoplacophoran genus Knightoconus upon the origin of Cephalopoda. Lethaia 6, 275–309 (1973)
Lindgren, A. R., Giribet, G. & Nishiguchi, M. K. A combined approach to the phylogeny of Cephalopoda (Mollusca). Cladistics 20, 454–486 (2004)
Strugnell, J. & Nishiguchi, M. K. Molecular phylogeny of coleoid cephalopods (Mollusca: Cephalopoda) inferred from three mitochondrial and six nuclear loci: a comparison of alignment, implied alignment and analysis methods. J. Molluscan Stud. 73, 399–410 (2007)
Runnegar, B. & Pojeta, J., Jr Molluscan phylogeny: the paleontological viewpoint. Science 186, 311–317 (1974)
Waller, T. R. in Bivalves: An Eon of Evolution (eds Johnston, P. A. & Haggart, J. W. ) 1–45 (Univ. Calgary Press, 1998)
Ponder, W. F. & Lindberg, D. R. Towards a phylogeny of gastropod molluscs: an analysis using morphological characters. Zool. J. Linn. Soc. 119, 83–265 (1997)
Aktipis, S. W. & Giribet, G. A phylogeny of Vetigastropoda and other ‘archaeogastropods’: re-organizing old gastropod clades. Invertebr. Biol. 129, 220–240 (2010)
Wilson, N. G. et al. Field collection of Laevipilina hyalina McLean, 1979 from southern California, the most accessible living monoplacophoran. J. Molluscan Stud. 75, 195–197 (2009)
Ewen-Campen, B. et al. The maternal and early embryonic transcriptome of the milkweed bug Oncopeltus fasciatus. BMC Genomics 12, 61 (2011)
Parkinson, J. et al. PartiGene: constructing partial genomes. Bioinformatics 20, 1398–1404 (2004)
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008)
Wasmuth, J. D. & Blaxter, M. L. prot4EST: translating expressed sequence tags from neglected genomes. BMC Bioinformatics 5, 187 (2004)
van Dongen, S. A Cluster Algorithm for Graphs Technical Report No. INS-R0010 (National Research Institute for Mathematics and Computer Science in the Netherlands, Amsterdam, 2000)
Apeltsin, L., Morris, J. H., Babbitt, P. C. & Ferrin, T. E. Improving the quality of protein similarity network clustering algorithms using the network edge weight distribution. Bioinformatics 27, 326–333 (2011)
Katoh, K. & Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics 9, 286–298 (2008)
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000)
Stamatakis, A. P. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006)
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008)
Whelan, S. & Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 (2001)
Huelsenbeck, J., Larget, B., van der Mark, P., Ronquist, F. & Simon, D. MrBayes: Bayesian Analysis of Phylogeny 〈http://mrbayes.csit.fsu.edu/index.php〉 (2005)
Lartillot, N., Lepage, T. & Blanquart, S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009)
Lartillot, N. & Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004)
Acknowledgements
This research was supported by the US National Science Foundation through the Systematics Program (awards 0844596, 0844881 and 0844652), the AToL Program (EF-0531757), EPSCoR (Infrastructure to Advance Life Sciences in the Ocean State, 1004057) and the iPlant Collaborative (0735191). Support was also provided by the Scripps Institution of Oceanography, the University of California Ship Funds and the Museum of Comparative Zoology. Collecting in Greenland was supported by the Carlsberg Foundation. A. Riesgo and J. Harasewych provided tissue samples of Octopus and Perotrochus, respectively. E. Röttinger assisted with Nautilus. C. Palacín allowed us to use an Octopus vulgaris photograph. At Brown University, Illumina sequencing was enabled by the Genomics Core Facility, and computational analyses were facilitated by L. Dong and the Center for Computing and Visualization. At Harvard University, Illumina sequencing was enabled by the Bauer Core in the Faculty of Arts and Sciences (FAS) Center for Systems Biology, and analyses were supported by the staff of the Research Computing cluster Odyssey facility in the FAS.
Author information
Authors and Affiliations
Contributions
C.W.D., G.G. and N.G.W. conceived of and oversaw the study. S.A.S. and C.W.D. designed and implemented the data analyses. N.G.W., G.G. and G.W.R. collected the specimens. F.E.G., S.C.S.A. and C.F. prepared the specimens for sequencing. S.A.S., C.W.D., G.G., G.W.R. and N.G.W. wrote the manuscript. All authors read and provided input into the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-9 with legends and Supplementary Table 1. This file contains updated versions of Supplementary Figures 2-9 and was replaced online on 28 November 2012 (see Corrigendum doi:10.1038/nature11736 for details). (PDF 489 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Smith, S., Wilson, N., Goetz, F. et al. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364–367 (2011). https://doi.org/10.1038/nature10526
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10526
This article is cited by
-
A chromosome-level genome assembly of a deep-sea symbiotic Aplacophora mollusc Chaetoderma sp.
Scientific Data (2024)
-
Shell field morphogenesis in the polyplacophoran mollusk Acanthochitona rubrolineata
EvoDevo (2023)
-
Comparative proteomics of the shell matrix proteins of Nautilus pompilius and the conchiferans provide insights into mollusk shell evolution at the molecular level
Marine Biology (2023)
-
Elemental analyses reveal distinct mineralization patterns in radular teeth of various molluscan taxa
Scientific Reports (2022)
-
Mito-nuclear coevolution and phylogenetic artifacts: the case of bivalve mollusks
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.