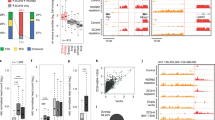
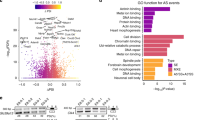
Abstract
Alternative splicing of pre-messenger RNA is a key feature of transcriptome expansion in eukaryotic cells, yet its regulation is poorly understood. Spliceosome assembly occurs co-transcriptionally, raising the possibility that DNA structure may directly influence alternative splicing. Supporting such an association, recent reports have identified distinct histone methylation patterns, elevated nucleosome occupancy and enriched DNA methylation at exons relative to introns. Moreover, the rate of transcription elongation has been linked to alternative splicing. Here we provide the first evidence that a DNA-binding protein, CCCTC-binding factor (CTCF), can promote inclusion of weak upstream exons by mediating local RNA polymerase II pausing both in a mammalian model system for alternative splicing, CD45, and genome-wide. We further show that CTCF binding to CD45 exon 5 is inhibited by DNA methylation, leading to reciprocal effects on exon 5 inclusion. These findings provide a mechanistic basis for developmental regulation of splicing outcome through heritable epigenetic marks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
03 November 2011
Panel labelling in Fig. 3e was corrected.
References
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008)
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 40, 1413–1415 (2008)
Tazi, J., Bakkour, N. & Stamm, S. Alternative splicing and disease. Biochim. Biophys. Acta 1792, 14–26 (2009)
Matlin, A. J., Clark, F. & Smith, C. W. Understanding alternative splicing: towards a cellular code. Nature Rev. Mol. Cell Biol. 6, 386–398 (2005)
Han, J., Xiong, J., Wang, D. & Fu, X. D. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 21, 336–343 (2011)
Singh, R. & Valcarcel, J. Building specificity with nonspecific RNA-binding proteins. Nature Struct. Mol. Biol. 12, 645–653 (2005)
Kornblihtt, A. R. Coupling transcription and alternative splicing. Adv. Exp. Med. Biol. 623, 175–189 (2007)
de la Mata, M. et al. A slow RNA polymerase II affects alternative splicing in vivo . Mol. Cell 12, 525–532 (2003)
Tilgner, H. et al. Nucleosome positioning as a determinant of exon recognition. Nature Struct. Mol. Biol. 16, 996–1001 (2009)
Spies, N., Nielsen, C. B., Padgett, R. A. & Burge, C. B. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell 36, 245–254 (2009)
Andersson, R., Enroth, S., Rada-Iglesias, A., Wadelius, C. & Komorowski, J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 19, 1732–1741 (2009)
Kolasinska-Zwierz, P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 41, 376–381 (2009)
Schwartz, S., Meshorer, E. & Ast, G. Chromatin organization marks exon-intron structure. Nature Struct. Mol. Biol. 16, 990–995 (2009)
Chodavarapu, R. K. et al. Relationship between nucleosome positioning and DNA methylation. Nature 466, 388–392 (2010)
Hodges, E. et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 19, 1593–1605 (2009)
Luco, R. F., Allo, M., Schor, I. E., Kornblihtt, A. R. & Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26 (2011)
Alló, M. et al. Chromatin and alternative splicing. Cold Spring Harb. Symp. Quant. Biol. 75, 103–111 (2010)
Trowbridge, I. S. & Thomas, M. L. CD45: an emerging role as a protein tyrosine phosphatase required for lymphocyte activation and development. Annu. Rev. Immunol. 12, 85–116 (1994)
Hermiston, M. L., Xu, Z. & Weiss, A. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 21, 107–137 (2003)
Oberdoerffer, S. et al. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 321, 686–691 (2008)
Topp, J. D., Jackson, J., Melton, A. A. & Lynch, K. W. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 14, 2038–2049 (2008)
Motta-Mena, L. B., Heyd, F. & Lynch, K. W. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol. Cell 37, 223–234 (2010)
Tong, A., Nguyen, J. & Lynch, K. W. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J. Biol. Chem. 280, 38297–38304 (2005)
Horgan, K. J. et al. CD45RB expression defines two interconvertible subsets of human CD4+ T cells with memory function. Eur. J. Immunol. 24, 1240–1243 (1994)
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007)
UCSC Genome Browser, GRC37/hg19, ENCODE Histone Modification Tracks.
Phillips, J. E. & Corces, V. G. CTCF: master weaver of the genome. Cell 137, 1194–1211 (2009)
Ohlsson, R., Bartkuhn, M. & Renkawitz, R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma 119, 351–360 (2010)
Luco, R. F. et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010)
Wada, Y. et al. A wave of nascent transcription on activated human genes. Proc. Natl Acad. Sci. USA 106, 18357–18361 (2009)
Buratowski, S. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36, 541–546 (2009)
Komissarova, N., Kireeva, M. L., Becker, J., Sidorenkov, I. & Kashlev, M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 371, 233–251 (2003)
Filippova, G. N. et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16, 2802–2813 (1996)
Renda, M. et al. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 282, 33336–33345 (2007)
Kim, T. H. et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 (2007)
Lyko, F. et al. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8, e1000506 (2010)
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007)
Jothi, R., Cuddapah, S., Barski, A., Cui, K. & Zhao, K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 36, 5221–5231 (2008)
Katz, Y., Wang, E. T., Airoldi, E. M. & Burge, C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature Methods 7, 1009–1015 (2010)
Huff, J. T., Plocik, A. M., Guthrie, C. & Yamamoto, K. R. Reciprocal intronic and exonic histone modification regions in humans. Nature Struct. Mol. Biol. 17, 1495–1499 (2010)
Berdasco, M. & Esteller, M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev. Cell 19, 698–711 (2010)
David, C. J. & Manley, J. L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 (2010)
Jelinic, P. & Shaw, P. Loss of imprinting and cancer. J. Pathol. 211, 261–268 (2007)
Mohn, F., Weber, M., Schubeler, D. & Roloff, T. C. Methylated DNA immunoprecipitation (MeDIP). Methods Mol. Biol. 507, 55–64 (2009)
Kireeva, M. L. et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell 30, 557–566 (2008)
Awrey, D. E. et al. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 272, 14747–14754 (1997)
Kireeva, M. L., Lubkowska, L., Komissarova, N. & Kashlev, M. Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae . Methods Enzymol. 370, 138–155 (2003)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009)
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009)
Ramsköld, D., Wang, E. T., Burge, C. B. & Sandberg, R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 5, e1000598 (2009)
Wall, M. E., Dyck, P. A. & Brettin, T. S. SVDMAN–singular value decomposition analysis of microarray data. Bioinformatics 17, 566–568 (2001)
Wall, M. E., Rechsteiner, A. & Rocha, L. M. in A Practical Approach to Microarray Data Analysis (eds Berrar, D. P., Dubitzky, W. & Granzow, M.) pp. 91–109 (Springer, 2003)
Acknowledgements
We thank A. Rao, C. Burge and K. Lynch for critical reading of this manuscript. We also thank A. Rao for reagents and K. Nyswaner and M. Prigge for technical assistance. This work is supported by the Intramural Research Program of NIH, the National Cancer Institute, The Center for Cancer Research (S.O., P.O., M.K.), and the Swedish Research Council Foundation and the Foundation for Strategic Research (R.S.).
Author information
Authors and Affiliations
Contributions
S.S. performed ChIP, MedIP and EMSA. M.G. and S.S. performed lentiviral transductions, transfections, flow cytometry, and qPCR. E.K. analysed ChIP and RNA-seq data. M.I. performed in vitro transcription. S.S. and B.S. cloned the minigenes. All authors designed experiments and M.K., P.O., R.S. and S.O. supervised the project. S.O. and R.S. wrote the text. P.O. edited the text.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-10 with legends and Supplementary Tables 1-3. (PDF 1904 kb)
Supplementary Table 4
The table shows a list of exons with significantly different inclusion levels after CTCF knock-down. (XLS 1395 kb)
Rights and permissions
About this article
Cite this article
Shukla, S., Kavak, E., Gregory, M. et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 (2011). https://doi.org/10.1038/nature10442
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10442
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.