Abstract
Crohn’s disease (CD) is an inflammatory pathology of the mucosal intestine that results from uncontrolled immune response towards commensal microbes. Clonal expansions of T cells have been found in patients with CD suggesting an antigen-specific stimulation of pathogenic T cells. Here we show, using T-cell receptor repertoire analysis by real-time PCR, that oligoclonal expansions are found in both CD8+ and CD4+ T cells in the blood and intestinal mucosa of CD patients. The majority of CD4+ T-cell-expanded clones are CD4+NKG2D+ T cells. These clonal expansions were found in both inflamed and neighboring healthy tissue and were persisting during the course of the disease. The presence of these CD4+NKG2D+ T-cell clones at the macroscopically normal edge of the surgical resection might be predictive of inflammation relapse post surgery.
Similar content being viewed by others
Introduction
Crohn’s disease (CD) is an inflammatory bowel disease that—together with ulcerative colitis—is characterized by chronic and relapsing inflammation of intestinal segments.1 CD pathogenesis remains unclear but is associated with abnormalities in intestinal mucosal barrier integrity2 and function,3 gut microbiota,4 and immune regulation.5, 6 CD is viewed as predominantly T-cell driven, and chronic intestinal inflammation may correlate either to impaired regulatory T-cell activity or excessive effector T-cell function. The breakdown of this balance results in aberrant immune responses presumably directed against the intestinal microbiota.7, 8, 9
Defects in innate immunity in CD—revealed by human genome-wide association studies10, 11—are exemplified by the identification of CD-associated NOD2/CARD15 gene variants with attenuated function.3, 12, 13 Impaired innate immunity in CD could result in aberrant amount of normally cleared intestinal antigens (Ags) and induce the stimulation of pathogenic T cells.14 Consistent with recognition of microbial Ags, T helper type 1 (Th1)-mediated colitis in mice is completely abrogated under germ-free conditions.15 In human, increased oligoclonal expansions of T cells in the peripheral blood (PB) and the inflamed mucosa of CD patients indicate Ag-specific activation of pathogenic T cells.16 Consistently, serum responses to several bacterial components are found in CD patients.17
We previously showed that a subset of natural killer group 2, member D (NKG2D)-expressing CD4+ T cells is selectively increased in the lamina propria (LP) and PB of CD patients but not in ulcerative colitis patients.18 NKG2D is an activating receptor that delivers a co-stimulatory signal during T-cell receptor (TCR)-mediated Ag recognition.19 NKG2D ligands are upregulated on the epithelium in both CD and ulcerative colitis active lesions.18 In normal conditions, CD4+ T cells almost express no NKG2D that is usually expressed on CD8+ T cells, γδ T cells, and NK cells.20 CD4+NKG2D+ T cells are also increased in other inflammatory conditions, including rheumatoid arthritis,21 and in infectious disease, such as cytomegalovirus22 and mycobacterium23 infections. In CD patients, CD4+NKG2D+ T-cell subset exhibits cytotoxic and proinflammatory properties, expressing high amounts of perforin, interferon-γ, and tumor necrosis factor-α, presumably contributing to intestinal inflammation.18 More recently, we demonstrated that NKG2D is a functional marker of Th17 cells in CD and that costimulation through NKG2D induces Th17-like responses.24 CD4+ T cells expressing NKG2D produce high level of interleukin (IL)-17 and IL-22 in CD and highly express C-C motif chemokine receptor 6, IL-23 receptor, and CD161, as well as the Th17 transcription factor RORC (retinoid-related orphan receptor-γ). Moreover, IL-17-producing CD4+ T cells highly expressed both NKG2D and CD161. Specific stimulation of both NKG2D and TCR significantly increased interferon-γ, IL-17, and tumor necrosis factor-α production in CD4+ T cells as compared with TCR activation alone. Moreover, CD4+NKG2D+ T cells showed a high sensitivity to Th17 polarization.24
The antigenic experience of a T-cell population can be assessed by analyzing the TCR repertoire defined by the number and frequency of TCR recombinations among T cells. A useful approach is the evaluation of the length of the CDR3 hypervariable regions (CDR3β) of the TCR Vβ-chains (TCR Vβ variable (TRBV)) that contacts the major histocompatibility complex molecules and their bound antigenic peptides.25, 26 By this method, Ag-specific expansions of T-cell clones—expressing the same appropriate TCR as their progenitor—are detected as a predominant CDR3β of a particular length. Within a given CDR3β spectrum, a predominant CDR3β does not necessarily indicate a monoclonal T-cell expansion and may represent a limited number of distinct T-cell clones expressing TCRs with the same CDR3β length (oligoclonal expansions). In contrast, Gaussian distributions of CDR3β segments indicate polyclonality. Consistent with the constant state of T-cell activation in the mucosa, oligoclonal expansions of T cells are found in the normal intestine.27, 28 Importantly, CD patients present decreased diversity of the TCR repertoire than controls.16 Previous work have shown the expansions of CD4+ T-cell clones in the inflamed tissues of CD patients;27, 28, 29 however, no attempt was made to answer the questions of the persistence of these expansions over time, more specifically after surgical treatment.
In this study, we combined qualitative and quantitative methods to precisely identify and quantify oligoclonal expansions of T cells with a superior accuracy than in previous studies. We investigated the TCR repertoire of several peripheral and mucosal CD8+ or CD4+ T cells in CD patients. Following our previous work on the increased numbers of CD4+NKG2D+ T cells in CD patients, we show that the majority of the CD4+ T-cell expansions occur in this specific subset. The CD4+NKG2D+ T-cell clonal expansions seen in the surgical specimen were found in the inflamed tissue during disease relapse. In a small cohort of patients, we show that the presence of these CD4+NKG2D+ T-cell clones at the non-inflamed edge of the surgical specimen might be predictive of disease relapse at 6 months post surgery.
Results
Increased T-cell oligoclonal expansions in Crohn’s disease
We used quantitative techniques of RNA amplifications to determine the TRBV chain usage in T cells (see Supplementary Figure S1 online) from CD and control patients included in the two cohorts (Table 1). By combining the identification of the TRBV segment with the determination of the length of the hypervariable region of this chain (CDR3β), we found oligoclonal expansions in PB CD8+ and CD4+ T cells from CD patients, as shown for one representative patient in Figure 1a. We quantified this “oligoclonality” by expressing the percentage represented by these clonal expansions reported to the usage of all TRBV (Figure 1a). The numbers obtained for each CD patients of the two cohorts showed that oligoclonality was increased in PB CD8+ T cells of CD patients compared with controls (Figure 1b). A similar trend was observed in CD4+ T cells (Figure 1c). Interestingly, similar results were obtained when T cells were isolated from the intestinal LP (Figure 1b).
Increased CD4+ and CD8+ T-cell oligoclonal expansions in Crohn’s disease (CD). (a) Representative immunoscope study of the T-cell receptor repertoire in peripheral blood (PB) CD8+ and CD4+ T cells from a CD patient. The values of the CDR3β length in amino acid (aa) for each TCR beta variable (TRBV)-chain according to its relative expression among the 25 analyzed TRBV families were plotted in 3D graphics. Black cones indicate oligoclonal expansions. The percentage of oligoclonality was calculated as indicated in the Methods section. Sorted (b) CD8+ and (c) CD4+ T-cell subsets were analyzed from CD patients (n=9 and n=5, respectively) and controls (Ctrl; n=4) in the PB or intestinal lamina propria (LP). The Mann–Whitney test was used for statistical analyses (*P<0.05).
CD4+NKG2D+ T cells contain most CD4+ oligoclonal expansions, which differ in the blood and intestinal mucosa
We recently demonstrated that a subset of CD4+ T cells expands in CD. These T cells express the NK receptor NKG2D and display specific features that may contribute to the pathology.18, 24 We hypothesized that the increased number of CD4+NKG2D+ T cells was accompanied by an expansion that could be visualized by our immunoscope methodology. Indeed, sorted CD4+NKG2D+ T cells from both the PB and LP showed significant increase of clonal expansion compared with CD4+NKG2D− cells (Figure 2a–c). In fact, CD4+NKG2D+ T cells encompass most of these expanded populations, confirming our hypothesis.
CD4+NKG2D+ T cells encompass most oligoclonal expansions found in CD4+ T cells from the intestinal mucosa of Crohn’s disease (CD) patients. (a) CD4+NKG2D+ (left) and CD4+NKG2D− (right) T-cell subsets were sorted from the peripheral blood (PB) of one representative patient (CD#3′) and T-cell receptor (TCR) repertoire patterns analyzed as in Figure 1. (b) T cells from the intestinal lamina propria (LP) analyzed as in panel a. CDR3β-specific oligoclonal expansions of CD4+NKG2D+ (black peaks) and CD4+NKG2D− (white peaks) LP T cells are shown. (c) TCR repertoire oligoclanility rate in CD4+NKG2D+ and CD4+NKG2D− in PB and LP of CD patients (n=7). NKG2D, natural killer group 2, member D; TRBV, TCR Vβ variable. *P<0.05.
We compared the oligoclonal expansions in CD4+NKG2D+ T cells from the PB and LP of the same patients. Surprisingly, the TRBV usage and CDR3β length of clonal expansions in these two compartments were different, suggesting that the clonal expansions seen in the LP are the consequence of local accumulation (Figure 2a).
Oligoclonal expansions with identical TRBV usage are found inside and outside the ileal inflammatory site
We next aimed at comparing the clonality of the T-cell populations found at the inflamed and the non-inflamed areas of the intestinal mucosa, the latter corresponding to the edge of the surgical specimen. Several clonal expansions visible in total mucosal T cells from the inflamed area were also found in the non-inflamed area (Figure 3a). Then, we sorted CD4+NKG2D+, CD4+NKG2D−, and CD8+ LP T cells and matched oligoclonal expansions to attribute a phenotype to each clone. Peaks that were found in total T cells were mainly observed in CD4+NKG2D+ and CD8+ T cells (Figure 3a). None of the peaks found in the CD4+NKG2D+ subset were present in CD4+NKG2D− T cells (data not shown). Thus, we were able to confirm that oligoclonal expansions were predominant in CD4+NKG2D+ T cells, and many oligoclonal expansions in this subset were found in both the inflamed and non-inflamed tissue (Figure 3c). These results demonstrate that reactive CD4+NKG2D+ and CD8+ clones of T cells are present beyond the inflamed area in the neighboring tissue.
Oligoclonal expansions are found inside and outside the ileal inflammatory site. (a) T-cell receptor (TCR) repertoire patterns of total T cells isolated from the inflamed and non-inflamed areas in one representative Crohn’s disease (CD) patient. (b) Sorted CD4+NKG2D+ and CD8+ lamina propria (LP) T cells from inflamed area were compared with the TCR repertoire from the same patient as in a. (c) Oligoclonality rates of total oligoclonal expansions related to sorted CD4+NKG2D−, CD4+NKG2D+, and CD8+ LP T cells, or unrelated within biopsies taken in macroscopically inflamed (left) and non-inflamed (right) areas from CD patients (n=5). The Mann–Whitney test was used for statistical analyses (*P<0.05). (d) Relative proportion (%) of CD4+NKG2D−, CD4+NKG2D+, CD8+ LP T cells, and unrelated oligoclonal expansions in the total oligoclonal expansions found in biopsies from macroscopically inflamed (left) and non-inflamed (right) areas. NKG2D, natural killer group 2, member D; TRBV, TCR Vβ variable.
Persistence of oligoclonal expansions of mucosal CD4+NKG2D+ T cells in post-operative relapse
We investigated the possibility that one of these reactive CD4+NKG2D+ T-cell clone might remain present in the tissue after surgery and during inflammation relapse. T-cell repertoire analysis was performed at several time points after surgery for one patient. The surgical procedure removed all inflammatory lesions located at the terminal ileum. An important expansion of CD4+NKG2D+ T cells using a TRBV5-2 segment with a eight amino-acid long CDR3β was found in both the inflamed and non-inflamed areas of this specimen (Figure 4a). Six months after surgery, the systematic endoscopic control revealed multiple ulcerations on the neo-terminal ileum beyond the ileocolic anastomosis. The same CD4+NKG2D+ T cells expansion using a TRBV5-2 segment was observed in the inflamed tissue biopsies (Figure 4b). The patient experienced a clinical relapse during the course of his treatment and, again, the same expansion was detected in the inflamed ileum and ileocecal anastomosis at 24 months post surgery (Figure 4c). In contrast, this specific Vβ segment was not found in the mucosa of the right colon and rectum (Figure 4e).
Persistence of CD4+NKG2D+ T-cell clones in the inflamed intestinal mucosa of Crohn’s disease (CD) patients experiencing endoscopic relapse. The T-cell receptor (TCR) repertoire patterns of one patient (CD#3′) are presented for total (unsorted) T cells from biopsies taken within macroscopically inflamed areas at (a) M0 and at the time of endoscopic recurrence (b) (M6). At 24 months post surgery (M24), biopsies were taken not only within macroscopically inflamed areas of (c) the ileum and (d) ileocecal anastomosis, but also at distance, (e) in the healthy right colon and rectum. In parallel, T-cell subset were sorted and oligoclonal expansions were matched according to their phenotype: CD4+NKG2D− (grey peaks), CD4+NKG2D+ (black peaks), CD8+ LP T cells (white peaks). NKG2D, natural killer group 2, member D; TRBV, TCR Vβ variable.
To further assess the clonality of this CD4+NKG2D+ T-cell expansion, products from the TRBV5-2 C2 PCR of the immunoscope were cloned and then sequenced. One single sequence was identified in the majority of the clones (72%; Figure 5). The exact same sequence corresponded to the major expansion at 6 months post surgery in the inflamed tissue (Figure 5). Interestingly, different clones with close amino-acid sequences were also identified at both time points, including two with an identical amino-acid CDR3β sequence (Figure 5) suggesting an Ag-driven process. Taken together, these results show that the same clonal expansions are present in the inflamed tissues at different time of the disease despite the treatments and surgical procedure undergone by the patient. These specific clones could have a major role in disease relapse.
Sequence analyses of persistent CD4+NKG2D+ T-cell clones in the inflamed intestinal mucosa at M0 (surgery) and M6 (endoscopic relapse) in one CD patient (CD#3′). Products from the TRBV5-2 C2 PCR of the immunoscope were cloned and then sequenced. The proportion of identical sequence obtained from different clones is indicated as a percentage. The TCR Vβ variable (TRBV), J, and C family names and the amino-acid sequences of the CDR3β segments are shown. NKG2D, natural killer group 2, member D.
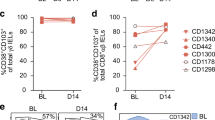
Presence of CD4+NKG2D+ T-cell clones at the ileal edge of the surgical specimen may predict disease relapse
We correlated the presence of these CD4+NKG2D+ oligoclonal expansions in the non-inflamed tissue at the time of surgery and the potential relapse at 6 months after the procedure. We followed five patients after surgery. We identified oligoclonal expansions in the inflamed and non-inflamed areas and analyzed their presence in sorted CD4+NKG2D+, CD4+NKG2D−, and CD8+ T-cell subpopulations. Inflamed and non-inflamed areas shared clonal expansions in four out of the five patients (Table 2). In these patients, we confirmed that most of these clones were CD4+NKG2D+ cells (Table 2). All three patients who had shared CD4+NKG2D+ clonal expansions in the inflamed tissue and on the edge of the surgical specimen had a relapse with ulcerations in the neo-terminal ileum 6 months after surgery while the two who did not stayed disease free for this period. These results suggest that the presence of CD4+NKG2D+ clones in the non-inflamed tissue at the edge of the surgical specimen may predict disease relapse after surgery.
Discussion
Increased T-cell oligoclonal expansions in the inflamed mucosa of CD patients suggest the recognition of a limited number of Ags.7, 16 In this work, we demonstrate that most of these oligoclonal expansions occur in CD4+ T cells, essentially in the subset expressing NKG2D. Despite the fact that they represent a small fraction of mucosal T cells, CD4+NKG2D+ T cells predominantly contribute to the global T-cell oligoclonality in the mucosa, even compared with mucosal CD8+ T cells that are approximately five times more numerous. As previously reported, there was no common expansion among patients supporting the absence of shared and dominant antigenic response in CD.27, 28, 29
Our data suggest that oligoclonally expanded CD4+ T cells are crucial actors in the persistent uncontrolled inflammation in CD. First, the expression of NKG2D identifies CD4+ T cells that exhibit pro-inflammatory and effector properties in CD.18, 24 Second, predominant oligoclonal expansions of LP CD4+NKG2D+ T cells were found within recurrent endoscopic lesions 6 months after surgery, suggesting that the persistence of LP CD4+NKG2D+ T-cell clones is associated with endoscopic and clinical relapse. In addition, these CD4+NKG2D+ T-cell clones appeared to be limited to lesional areas and their immediate surroundings (ileal edge of the surgical specimen) but not in distant and non-involved mucosa of the colon or rectum, as observed in one patient.
Currently, no early predictive factor of disease relapse has been validated, and immunosuppressive treatment is usually proposed in case of significant endoscopic lesions investigated 6 months after surgery.30 Recent studies have shown that early introduction of anti-tumor necrosis factor monoclonal antibodies after surgery dramatically decrease endoscopic relapses.31, 32, 33 Thus, a predictive marker is needed to identify patients with high risks of endoscopic recurrence who may benefit from early post-operative treatments. In this study, we observed a diffusion of T-cell clones in macroscopically normal mucosal areas located at the ileal edge of the surgical specimen. This suggests that pathogenic T cells might still be present—after surgical removal of lesional tissues—in adjacent non-resected macroscopically normal mucosa forming the ileocecal anastomosis. Indeed, the dissemination of strongly expanded CD4+NKG2D+ T-cell clones to the ileal edge of the surgical specimen was associated with post-operative endoscopic recurrence at 6 months. These results, which should be confirmed in a larger prospective study, suggest that the persistence of pathogenic CD4+NKG2D+ T-cell clones in the neo-terminal ileum may have a role in post-operative recurrence.
Interestingly, the CD4+NKG2D+ T cell oligoclonal expansions from the LP were not found as such in the PB. The accumulation of specific clones in the LP might be related to local factors inducing survival and/or cell cycling, which are absent in the PB. Such signals could stem from soluble molecules like cytokines, such as IL-15.18 Additionally, cellular interactions could be involved in this process. Intestinal epithelial cells from CD patients express NKG2D ligands, which induce the costimulation and expansion of CD4+NKG2D+ T cells in vitro.18 Ag specificity is most likely involved in the accumulation of these clonal expansions in the mucosal tissues. It would be of interest to test the specificity of these CD4+NKG2D+ T cells towards microbial or environmental Ags and if this recognition induces a proliferation in situ.
Most studies of the TCR repertoire in CD focused on CD4+ T cells rather than CD8+ T cells. It was demonstrated that CD4+ T cells had the most distinct TCR repertoires between the LP and the PB in CD patients.34 Moreover, the basal oligoclonality of CD8+ T cells in controls questioned the disease-specificity of CD8+ T cells in CD. Recently, mice models of hapten-induced colitis reinstated the potent involvement of Ag-specific CD8+ T cells in CD.35 In this study, we show that the oligoclonality rates of PB and LP CD8+ T cells were significantly superior in CD patients as compared with controls, supporting the presence of Ag-driven activation of CD8+ T cells in CD. As CD8+ T cells constitutively express NKG2D—through which they exert cytotoxic activity in celiac disease36—CD8+ and CD4+NKG2D+ T cells may share activation mechanisms. However, conversely to CD4+NKG2D+ T cells, some oligoclonal expansions of CD8+ T cells were observed in both the LP and PB of half the patients, as well as in distant and non-involved mucosa of the colon or rectum, suggesting that some CD8+ T-cell clones may not be disease-specific. A better characterization of effector CD8+ T cells in CD is crucial to help distinguish pathogenic from non-pathogenic populations.
In conclusion, we show that most T-cell oligoclonal expansions found in the inflamed mucosa of CD patients correspond to CD4+ T cells expressing NKG2D. These pathogenic T-cell clones expand through Ag interactions. Anatomic diffusion and persistence of mucosal CD4+NKG2D+ T-cell clones delineate their implication in post-operative recurrence after surgery. Treatments aiming at the eradication of these pathogenic T-cell clones could decrease the risk of post-operative relapse and, more globally, could modify the outcome of the disease.
Methods
CD patients and controls. Surgical specimens from 21 patients with active CD undergoing bowel resection at Mount Sinai Medical Center (cohort 1, n=9) and at Hospital Saint-Louis (cohort 2, n=12) were used as the source for LP lymphocytes (LPLs). The group termed “controls” consisted of patients (n=7) undergoing bowel resection for diverticulitis or colorectal cancer. Clinical characteristics are given in Table 1. In the control group, LPLs were isolated from inflamed tissue (diverticulitis) or normal tissue located at least 10 cm away from tumor. In CD patients, LPL were isolated from the inflamed areas of resected specimens. Additionally in cohort 2, biopsies were taken within the macroscopically inflamed area and within macroscopically normal area at the ileal edge of the surgical specimens. All patients from cohort 2 underwent ileocecal resections. The ethical committees of Mount Sinai Medical Centre and Hospital Saint-Louis approved this study, and all subjects gave written informed consent.
Lymphocytes isolation and sorting of specific subsets. LPL isolation from surgical specimens was performed as previously described.37 PB lymphocytes were isolated from heparinized venous blood collected from controls and CD patients on a Ficoll-hypaque density gradient. LPL and PB lymphocytes were re-suspended in phosphate-buffered saline for cell sorting. In cohort 1, CD8+ T cells were sorted using the EasySep Human CD8-positive selection kit (StemCell Technologies, Grenoble, France) and the purity (>98%) was controlled by flow cytometry. In cohort 2, CD8+, CD4+, CD4+NKG2D+, and CD4+NKG2D− T cells were sorted using the Cell Sorter Vintage SE DIVA (Becton Dickinson, Le Pont de Claix, France) with mean purity >95%. For human cord blood CD4 T-cell separation, magnetic positive selection was performed as described by the manufacturer (Miltenyi Biotec, Paris, France) and the purity (>98%) was controlled by flow cytometry.
TRBV-specific CDR3 usage and length analysis. RNAs from isolated cells (10,000–200,000 cells) were reverse-transcribed into cDNA using Superscript III first-strand synthesis system for quantitative PCR (RT-qPCR) (Invitrogen, Saint-Aubin, France). An aliquot of cDNA synthesis reaction was amplified with each of 25 TRBV family-specific primers, together with a TRB constant region (TRBC) primer and a TaqMan (Saint-Aubin, France) probe for TRBC. CD3 primers and probes were used as internal controls. TRBV-specific and TRBC-specific primers have been described previously,38 except for some that were modified.
For immunoscope experiments, 2 μl of each of the amplification reactions described above were used as template in a run-off reaction initiated by a nested TRBC fluorescent primer in a total volume of 10 μl as described.39 The run-off reactions were analyzed on a 3100-Avant Genetic Analyzer (Applied Biosystems, Saint-Aubin, France).
Technical steps are summarized in Supplementary Figure S1 available online and have been tested on a small number of cells isolated from polyclonal cord blood CD4+ T cells (see Supplementary Figure S2 online).
TCR repertoire analysis: definitions and statistics. Combination of the relative representation of each CDR3β segments (immunoscope) with the relative expression of the relevant TRBV-chain among all the 25 analyzed TRBV-chain families (RT-qPCR) represented the relative usage of a given CDR3β segment among all the analyzed TCRs. These values were plotted in 3D graphics illustrating the relative percentage of the analyzed T cells expressing TCRs with distinct TRBV-chain and CDR3β size. TCRs with distinct TRBV-chain and CDR3β size were expressed as TRBVx(y) with x representing the primer-specific TRBV-chain family40 and y the length in amino acids (aa) of CDR3β segments.
The Shapiro–Wilk41 goodness-of-fit test was used to determine the normality of distribution of CDR3β segments within each TRBV family. Gaussian distribution (W>0.92) indicated polyclonality, whereas oligoclonal expansions of T cells were identified as the major peak in non-Gaussian distributions (W<0.75, P<0.01).
Oligoclonality rates were calculated by combining the relative area of the oligoclonal peak among all the detected peaks in non-Gaussian distributions of CDR3β segments in a given TRBV family and the relative expression of the 25 analyzed TRBV families.
Comparisons of oligoclonality rates between samples were performed using Wilcoxon–Mann–Whitney and t-tests (ANOVA). A P-value of <0.05 was considered to indicate statistical significance.
Among a given TRBV family, an oligoclonal expansion was identified as the major peak in non-Gaussian distributions of CDR3β segments. Oligoclonality rates, defined by the percentage of all oligoclonally expanded T cells (sum of all oligoclonal expansions) among analyzed T cells within a sample, were calculated by combining the relative area of the oligoclonal peak (immunoscope) and the relative expression of the relevant TRBV family (RT-qPCR).
TRBV cloning and sequencing. Amplified products were cloned using the TA cloning kit from Invitrogen. Plasmid DNA was prepared from single isolated colonies using the QIAamp DNA Mini Kit (Qiagen, Courtaboeuf, France), used for sequencing using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) on a 3130xl Genetic Analyzer (Applied Biosystems). Analyses were performed with Analysis software version 5.2 (Applied Biosystems), Bioedit software (Raleigh, NC), and Dambe software (Ottawa, ON, Canada).
The amplified CDR3β length was defined as the region between the upstream amino-acid CASS residues in the TRBV segments and the downstream GXG box in the TRBJ region.42
References
Podolsky, D.K. Inflammatory bowel disease. N. Engl. J. Med. 347, 417–429 (2002).
Buhner, S. et al. Genetic basis for increased intestinal permeability in families with Crohn's disease: role of CARD15 3020insC mutation? Gut 55, 342–347 (2006).
Wehkamp, J. et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc. Natl. Acad. Sci. USA 102, 18129–18134 (2005).
Darfeuille-Michaud, A. et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127, 412–421 (2004).
Xavier, R.J. & Podolsky, D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434 (2007).
Cobrin, G.M. & Abreu, M.T. Defects in mucosal immunity leading to Crohn's disease. Immunol. Rev. 206, 277–295 (2005).
Mowat, A.M. Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 3, 331–341 (2003).
Allez, M. & Mayer, L. Regulatory T cells: peace keepers in the gut. Inflamm Bowel Dis. 10, 666–676 (2004).
Bouma, G. & Strober, W. The immunological and genetic basis of inflammatory bowel disease. Nat. Rev. Immunol. 3, 521–533 (2003).
Barrett, J.C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
Consortium WTCC.. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Hugot, J.P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Hisamatsu, T. et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology 124, 993–1000 (2003).
Smith, A.M. et al. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn's disease. J. Exp. Med. 206, 1883–1897 (2009).
Elson, C.O. et al. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol. Rev. 206, 260–276 (2005).
Probert, C.S. et al. Repertoire of the alpha beta T cell receptor in the intestine. Immunol. Rev. 215, 215–225 (2007).
Ferrante, M. et al. New serological markers in inflammatory bowel disease are associated with complicated disease behaviour. Gut 56, 1394–1403 (2007).
Allez, M. et al. CD4+NKG2D+ T cells in Crohn's disease mediate inflammatory and cytotoxic responses through MICA interactions. Gastroenterology 132, 2346–2358 (2007).
Groh, V. et al. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat. Immunol. 2, 255–260 (2001).
Raulet, D.H. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 3, 781–790 (2003).
Groh, V. et al. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl. Acad. Sci. U S A 100, 9452–9457 (2003).
Saez-Borderias, A. et al. Expression and function of NKG2D in CD4+ T cells specific for human cytomegalovirus. Eur. J. Immunol. 36, 3198–3206 (2006).
Vankayalapati, R. et al. Role of NK cell activating receptors and their ligands in the lysis of mononuclear phagocytes infected with an intracellular bacterium. J. Immunol. 175, 4611–4617 (2005).
Pariente, B. et al. Activation of the receptor NKG2D leads to production of Th17 cytokines in CD4+ T cells of patients with Crohn's disease. Gastroenterology 141, 217–226 226.e1-2 (2011).
Jorgensen, J.L. et al. Mapping T cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature 355, 224–230 (1992).
Garboczi, D.N. et al. Structure of the complex between human T cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996).
Gulwani-Akolkar, B. et al. CD4+ cell oligoclonality in Crohn's disease: evidence for an antigen-specific response. Hum. Immunol. 48, 114–124 (1996).
Gulwani-Akolkar, B. et al. Selective expansion of specific T cell receptors in the inflamed colon of Crohn's disease. J. Clin. Invest. 98, 1344–1354 (1996).
Gulwani-Akolkar, B. et al. Crohn's disease is accompanied by changes in the CD4+, but not CD8+, T cell receptor BV repertoire of lamina propria lymphocytes. Clin. Immunol. Immunopathol. 77, 95–106 (1995).
ECCO. Guidelines dans JCC.
Sorrentino, D., Paviotti, A., Terrosu, G., Avellini, C., Geraci, M. & Zarifi, D. Low-dose maintenance therapy with infliximab prevents postsurgical recurrence of Crohn's disease. Clin. Gastroenterol. Hepatol. 8, 591–599 (2010).
Regueiro, M. et al. Infliximab prevents Crohn's disease recurrence after ileal resection. Gastroenterology 136, 441–450 (2009).
Papamichael, K., Archavlis, E., Lariou, C. & Mantzaris, G.J. Adalimumab for the prevention and/or treatment of post-operative recurrence of Crohn's disease: a prospective, two-year, single center, pilot study. J. Crohns. Colitis. 6, 924–931 (2012).
Blumberg, R.S. et al. Human intestinal intraepithelial lymphocytes are derived from a limited number of T cell clones that utilize multiple V beta T cell receptor genes. J. Immunol. 150, 5144–5153 (1993).
Nancey, S. et al. CD8+ cytotoxic T cells induce relapsing colitis in normal mice. Gastroenterology 131, 485–496 (2006).
Meresse, B. et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 21, 357–366 (2004).
Allez, M. et al. Expansion of CD8+ T cells with regulatory function after interaction with intestinal epithelial cells. Gastroenterology 123, 1516–1526 (2002).
Lim, A. et al. Combination of MHC-peptide multimer-based T cell sorting with the Immunoscope permits sensitive ex vivo quantitation and follow-up of human CD8+ T cell immune responses. J. Immunol. Methods 261, 177–194 (2002).
Clave, E. et al. Acute graft-versus-host disease transiently impairs thymic output in young patients after allogeneic hematopoietic stem cell transplantation. Blood 113, 6477–6484 (2009).
Wei, S. et al. The extent of the human germline T cell receptor V beta gene segment repertoire. Immunogenetics 40, 27–36 (1994).
Shapiro, S.S. & Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 52, 591–611 (1965).
Toyonaga, B. et al. Organization and sequences of the diversity, joining, and constant region genes of the human T cell receptor beta chain. Proc. Natl. Acad. Sci. USA 82, 8624–8628 (1985).
Acknowledgements
We thank Emeline Dufresne for technical assistance, Michel Schmid (IFR105) for cell sorting, and their colleagues of the Departments of Surgery (Emile Sarfati, Pierre Cattan, Nicolas Munoz-Bongrand, Paul Smereka, and Saurabh Mehandru). We are grateful to the late Professor Marc Lémann for his contribution during the conduct of the study. This work was supported by grants from INSERM.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
MA has received honoraria from Novo Nordisk as consultant. The other authors declared no conflict of interest.
Additional information
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Supplementary information
Rights and permissions
About this article
Cite this article
Camus, M., Esses, S., Pariente, B. et al. Oligoclonal expansions of mucosal T cells in Crohn’s disease predominate in NKG2D-expressing CD4 T cells. Mucosal Immunol 7, 325–334 (2014). https://doi.org/10.1038/mi.2013.51
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2013.51
This article is cited by
-
Expanded TCRβ CDR3 clonotypes distinguish Crohn's disease and ulcerative colitis patients
Mucosal Immunology (2018)
-
Regulatory NK1.1−CD4+NKG2D+ subset induced by NKG2DL+ cells promotes tumor evasion in mice
Cancer Immunology, Immunotherapy (2018)
-
An approach to the immunophenotypic features of circulating CD4+NKG2D+ T cells in invasive cervical carcinoma
Journal of Biomedical Science (2015)
-
Immune Responses to Intestinal Microbes in Inflammatory Bowel Diseases
Current Allergy and Asthma Reports (2015)