Abstract
The middle-frequency sensorineural hearing loss (MFSNHL) is rare among hereditary non-syndromic hearing loss. To date, only three genes are reported to be associated with MFSNHL, including TECTA, EYA4 and COL11A2. In this report, we analyzed and explored the clinical audiological characteristics and the causative gene of a Chinese family named HG-Z087 with non-syndromic autosomal dominant inherited MFSNHL. Clinical audiological characteristics and inheritance pattern of a family were evaluated, and pedigree was drawn based on medical history investigation. Our results showed that the Chinese family was characterized by late onset, progressive, non-sydromic autosomal dominant MFSNHL. Targeted exome sequencing, conducted using DNA samples of an affected member in this family, revealed a novel heterozygous missense mutation c.1643C>G in exon 18 of EYA4, causing amino-acid (aa) substitution Arg for Thr at a conserved position aa-548. The p.T548R mutation related to hearing loss in the selected Chinese family was validated by Sanger sequencing. However, the mutation was absent in control group containing 100 DNA samples from normal Chinese families. In conclusion, we identified the pathogenic gene and found that the novel missense mutation c.1643C>G (p.T548R) in EYA4 might have caused autosomal dominant non-syndromic hearing impairment in the selected Chinese family.
Similar content being viewed by others
Introduction
Hearing loss is the most common sensory disorder in human. It can be caused by genetic and environmental factors, such as medical factor, environment exposure, injury and medicine. Among the hereditary hearing loss, 70% is non-syndromic, which is human monogenic inherited disease. Non-syndromic hearing loss mainly affects high frequencies. In contrast, the middle-frequency (500–2000 Hz) sensorineural hearing loss (MFSNHL) is rare. To date, 168 non-syndromic deafness loci have been identified. In addition, 50 recessive (DFNB), 30 dominant (DFNA), 4X-linked (DFNX) and 1Y-linked (DFNY) genes have been cloned (http://hereditaryhearingloss.org). Only three genes are associated with MFSNHL, including TECTA (DFNA8/12),1, 2 EYA4 (DFNA10)3 and COL11A2 (DFNA13).4, 5
Previously, the primary method implemented to identify disease causative genes was conventional strategy of positional cloning and linkage analysis. However, there are considerable limitations. Recent years, targeted exome sequencing technology has become a powerful and affordable mean as to exploring such genes. There are numerous studies that showed the sensitivity and accuracy of targeted exome sequencing approach in the identification of the causal genes of hereditary hearing loss.6, 7, 8, 9, 10
Here, we report the application of targeted exome capture to identify a novel missense mutation c.1643C>G (p.T548R) in EYA4 for MFSNHL. We identified this gene with only one affected individual in one six-generation Chinese MFSNHL family. Targeted exome sequencing revealed a novel heterozygous missense mutation c.1643C>G in exon18 of EYA4, causing the amino-acid mutation of Thr to Arg at a conserved position 548. The p.T548R substitution was consistent with hearing loss in this Chinese family according to the results from Sanger sequencing. In addition, the mutation was absent in 100 unrelated control DNA samples of Chinese origin.
Materials and methods
Family and clinical evaluation
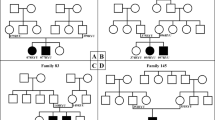
A Chinese family named HG-Z087 with late onset progressive sensorineural hearing loss was ascertained through the Department of Otolaryngology, Chinese PLA Wuhan General Hospital of Guangzhou Military Command. The pedigree of this family, spanning 6 generations and including 79 members, is consistent with an autosomal dominant inheritance pattern. Forty-four family members, including 18 affected and 26 unaffected individuals, participated in this study. The study was approved by the ethics committee of Chinese PLA Wuhan General Hospital of Guangzhou Military Command and appropriate informed consent was obtained from all participants accordingly. Medical history was obtained by using a questionnaire regarding the following aspects of this condition: subjective degree of hearing loss, age at onset, evolution, symmetry of the hearing impairment, use of hearing aids, the presence of tinnitus, pressure in the ears or vertigo, medication, noise exposure, pathological changes in the ear and other relevant clinical manifestations. Otoscopy, physical examination and pure tone audiometry (at frequencies from 250 to 8000 Hz) were performed to identify the phenotype. Acoustic immittance testing was applied to evaluate middle-ear pressures, ear canal volumes and tympanic membrane mobility. In two cases of the family (V-8 and V-9), distortion-product evoked otoacoustic emission and auditory brainstem response audiometry were measured, respectively. Computed tomography scan analysis of the proband (V-9) of the family has been conducted to rule out inner-ear malformations. All genomic DNA was extracted from peripheral blood using a blood DNA extraction kit according to the protocol provided by the manufacturer (QIAGEN, Hilden, Germany).
DNA library preparation
Each DNA sample is quantified by agarose gel electrophoresis and Nanodrop (Thermo-Fisher, Waltham, MA, USA). Libraries were prepared using Illumina standard protocol. Briefly, 3 μg of genomic DNA was fragmented by nebulization, the fragmented DNA is repaired, an ‘A’ is ligated to the 3′ end, Illumina adapters are then ligated to the fragments, and the sample is size selected aiming for a 350- to 400-base pair product. The size selected product is PCR amplified (each sample is tagged with a unique index during this procedure), and the final product is validated using the Agilent Bioanalyzer (Agilent, Santa Clare, CA, USA).
Disease genes enrichment and high-throughput sequencing
The amplified DNA was captured with a deafness disease-related Gene Panel using biotinylated oligo-probes (MyGenostics GenCap Enrichment Technologies, MyGenostics, Baltimore, MD, USA). The probes were designed to tile along 102 deafness system disease-related genes containing 3663 exons, 3 miRNA and 3 Mitochondrial genes relevant region (Supplementary Material,Supplementary Tables S1–S3). The capture experiment was conducted according to the manufacturer’s protocol. In brief, 1 μg DNA library was mixed with Buffer BL and GenCap gene panel probe (MyGenostics), heated at 95 °C for 7 min and 65 °C for 2 min on a PCR machine; 23 μl of the 65 °C prewarmed Buffer HY (MyGenostics) was then added to the mix, and the mixture was held at 65 °C with PCR lid heat on for 22 h for hybridization. In all, 50 μl MyOne beads (Life Technology) was washed in 500 μl 1X binding buffer for three times and resuspended in 80 μl 1X binding buffer. In all, 64 μl 2X binding buffer was added to the hybrid mix, and transferred to the tube with 80 μl MyOne beads. The mix was rotated for 1 h on a rotator. The beads were then washed with WB1 buffer at room temperature for 15 min once and WB3 buffer at 65 °C for 15 min three times. The bound DNA was then eluted with Buffer Elute. The eluted DNA was finally amplified for 15 cycles using the following program: 98 °C for 30 s (1 cycle); 98 °C for 25 s, 65 °C for 30 s, 72 °C for 30 s (15 cycles); 72 °C for 5 min (1 cycle). The PCR product was purified using SPRI beads (Beckman Coulter, Brea, CA, USA) according to the manufacturer’s protocol. The enrichment libraries were sequenced on Illumina HiSeq 2000 sequencer for paired read 100 bp.
Bioinformatics analysis
After HiSeq 2000 sequencing, high-quality reads were retrieved from raw reads by filtering out the low quality reads and adaptor sequences using the Solexa QA package and the cutadapt program (http://code.google.com/p/cutadapt/), respectively. SOAPaligner program was then used to align the clean read sequences to the human reference genome (hg19). After the PCR duplicates were removed by the Picard software, the single nucleotide polymorphisms (SNPs) were first identified using the SOAPsnp program (http://soap.genomics.org.cn/soapsnp.html). Subsequently, we realigned the reads to the reference genome using BWA and identified the insertions or deletions (InDels) using the GATK program (http://www.broadinstitute.org/gsa/wiki/index.php/Home_Page). The identified SNPs and InDels were annotated using the Exome-assistant program (http://122.228.158.106/exomeassistant). MagicViewer was used to view the short read alignment and validate the candidate SNPs and InDels. Non-synonymous variants were evaluated by four algorithms, Ployphen, SIFT, PANTHER and Pmut to determine pathogenicity.
Mutation analysis of the EYA4
The DNA from forty-four family members, 100 unrelated Chinese control individuals was analyzed by DNA sequencing. Primers were designed to amplify the18th exon and flanking intronic sequence of the EYA4 gene (NM_004100). The 18th exon and exon–intron boundaries of the EYA4 gene were amplified using standard PCR conditions and were completely sequenced. The primer sequences and precise PCR conditions are available from the authors on request. Bi-directional sequencing was carried out using both the forward and reverse primers, and was performed using the ABI PRISMBig Dye Terminator cycle sequencing ready reaction kit on a 3100 ABI DNA sequencer (Thermo-Fisher).
Results
Clinical feature of the family
The Chinese family HG-Z087 showed an autosomal dominant inheritance pattern (Figure 1). According to information collected from questionnaires, the age at onset of hearing impairment has varied from 17 to 40 years in this family. In all 18 affected subjects, the hearing impairment was symmetric while the severity varies significantly according to the audiograms, which could be classified into four different groups, including mild, moderate, severe and profound (Table 1). The hearing loss first affected the middle frequencies (<2000 Hz) and second involved all frequencies. Overall, the affected members have U-shaped or cookie-bite configuration. As shown in Figure 2, overlapping audiogram showed middle-frequency involved hearing loss with gradual progression.
Audiologic evaluation of the family members demonstrated normal acoustic immittance testing and bone conduction values that equal the air conduction measurements, suggesting sensorineural hearing impairment. No patients have a history of the use of amino-glycosides. A few affected members, including IV-28,V-2, V-8 and V-9, have a history of excessive exposure to noise. In two cases of the family (V-8 and V-9), distortion-product evoked otoacoustic emission and auditory brainstem response were measured respectively.
Computed tomography scan analysis of the proband (V-9) of the family has been conducted to rule out inner-ear malformations. Comprehensive family medical history and clinical examination of these individuals showed no other clinical abnormalities, including cardiovascular disease.
Candidate mutations identified by targeted exome sequencing
As shown in Table 2, 99.0% of the targeted disease gene regions were sequenced, and 95.1% of the targeted bases were sequenced with >10X depth that allowed to accurately call a SNP. The variants were functionally annotated using an in-house pipeline as well as the reported results from public available databases (dbSNP 135, HapMap database, 1000 genome variants database and a local control database) and categorized into different groups, including missense, non-sense, splice-site, insertion, deletion, synonymous and non-coding mutations. For all variants, the results were filtered with a quality value of single base sequencing ⩾20. A total number of 279 variants were identified in the sample. Among them, there were 163 non-synonymous variants, missense, non-sense and splicing variants. This was narrowed down to 6 by excluding variants reported in HapMap 28 and the SNP release of the 1000 Genome Project with minor allele frequency >0.01. For missense variants, computational prediction by PolyPhen, SIFT, PANTHER, PMUT as well as restriction such like consistency of genetic transmission mode further reduced the number of candidate mutations to 2. As a result, only one mutation potentially matched to the patient’s disease phenotype.
The identification of known pathogenic variants was based on mutations previously reported to cause MODY in the literature, in Locus-Specific Mutation Databases (LOVD, http://grenada.lumc.nl/LSDB_list/lsdbs) or previously identified to segregate with diabetes in Danish MODY families. Novel variants are only considered as pathogenic when they satisfied following criteria: (1) stop/frameshift variants; (2) missense mutations positioned in the amino-acid conserved region across species; (3) splice-site variations fulfilling the GT-AT rules; (4) predicted to be possibly damaging or disease-causing by more than two of the bioinformatic programs (SIFT, Sorting Intolerant From Tolerant, http://sift.bii.a-star.edu.sg/; PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/; Mutation Taster, http://www.mutationtaster.org/; BDGP, Berkeley Drosophila Genome Project, http://www.fruitfly.org/seq_tools/splice.html).
Mutation analysis of the EYA4
We sequenced the coding exons and about 100 bp of flanking intronic sequence of EYA4 from forty-four members and revealed a heterozygous missense mutation c.1643C>G in exon18 of EYA4 (NM_004100), causing amino-acid substitution Thr to Arg at a conserved position 548. The residue at 548 in EYA4 is highly conserved across Danio rerio, Xeponus, Mus musculus, Rattus, Gallus, Macaca, Homo sapiens and Bos taurus (Figure 3). Sequence analysis of 44 family members demonstrated that the c.1643C>G substitution was consistently correlated with hearing loss in this family, and that it was absent in 100 chromosomes of unrelated control subjects of Chinese origins. These findings together support the hypothesis that c.1643C>G represents a causative mutation, not just a rare polymorphism.
Mapping mutation analysis of EYA4 gene in Chinese family HG-Z087. (a) DNA sequence chromatogram shows the c. 1643C in wild type. (b) DNA sequence chromatogram shows the c.1643 C>G mutation in family HG-Z087. (c) Conservation analysis shows that T548R is highly conserved across Danio rerio, Xeponus, Mus musculus, Rattus, Gallus, Macaca, Homo sapiens and Bos.
Discussion
EYA4 is orthologous to the Drosophila gene eya and localized on 6q23.11 Its encoded protein contains a highly conserved carboxyl domain with 271 amino acid that was named eya-homologous region (eyaHR), and a more divergent proline-serine-threonine (PST)-rich (34–41%) transactivation domain at the N-terminus called eya variable region (eyaVR).11 The eyaHR is essential for the function among members of EYA protein family regarding their interactions with PAX, SIX and DACH proteins in the genetic network, which is conserved across species.12
In the present study, we report a Chinese family in which middle-frequency non-syndromic autosomal dominant hearing impairment segregates with the DFNA10 locus. By deafness genes panel enrichment (MyGenostics) and high-throughput sequencing, we identified a novel heterozygous missense mutation c.1643C>G in exon18 of EYA4, causing amino-acid mutation of Thr to Arg at a conserved position 548. Because the three-dimensional protein structure of EYA4 has not been available yet, we used eya2HR (PDB ID: 4egc.1) as the template to predict the likely structure impact of T548R substitution on EYA4 protein with PyMOL, based on the high similarity between human eya4HR and eya2HR (78.37% identity). The three-dimensional crystal structure of eyaHR domain of EYA4 (residues 369–639) and predicted the potential impacts of T548R substitution on EYA4 with PyMOL were shown in Figure 4. The modeled structure of mutated protein was composed of multiple-helixs, five β-sheets and several disordered loops. Specifically, five β-sheets are located inside of protein and close to the central domain. The residue T548 is located at the end of the second β-sheet. In the wild-type EYA4, there are three successive threonines (T548-T550) at this region. Threonine is a polar residue with the potential to form hydrogen bond interactions with other polar groups. In this case, the oxygen in the side chain of T548 forms a hydrogen bond with hydroxyl group of another T550. In contrast, in the R548 mutation, the interactions between residues are further strengthened by forming two new hydrogen bonds with hydroxyl groups of T549 and T550, respectively (Figure 4), which results in a conformational change in the protein structure.
Up to date, five families with MFSNHL at the DFNA10 locus have been reported. The genetic and clinical characteristics of all identified EYA4 mutations associated with the DFNA10 locus are summarized (Table 3). Among them, there are a non-sense mutation c. 2200C>T in a Belgian family,13, 14 one splicing mutation ca.1282-12T-A in an Australian family15 and three insertion mutations (1468insAA in an American family,14 1558insTTTG in a Hungarian family16 and 1490insAA in the second American family.17 All five EYA4 mutations detected in DFNA10 families resulted in premature termination codons. Five truncated mutations resulted in EYA4 eyaHR deletion (Belgium family, Hungary family and Australia family) or accompanied by partial deletion of eyaVR (two American families). On the basis of our work, a novel missense T548R mutation was identified in a Chinese family. The postlingual-onset, progressive MFSNHL phenotype segregating in Chinese family HG-Z087 is similar to what has been reported for DFNA10 families.
In conclusion, we identified a novel missense mutation of EYA4 in a Chinese DFNA10 family with MFSNHL through a deafness genes panel enrichment and high-throughput next-generation sequencing. Our data provide a better genotype–phenotype understanding about more than truncated mutation that could lead to DFNA10 phenotype.
References
Collin, R. W., de Heer, A. M., Oostrik, J., Pauw, R. J., Plantinga, R. F. & Huygen, P. L. et al. Mid-frequency DFNA8/12 hearing loss caused by a synonymous TECTA mutation that affects an exonic splice enhancer. Eur. J. Hum. Genet. 16, 1430–1436 (2008).
Verhoeven, K., Van Laer, L., Kirschhofer, K., Legan, P. K., Hughes, D. C. & Schatteman, I. et al. Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat. Genet. 19, 60–62 (1998).
De Leenheer, E. M., Huygen, P. L., Wayne, S., Verstreken, M., Declau, F. & Van Camp, G. et al. DFNA10/EYA4—the clinical picture. Adv. Otorhinolaryngol. 61, 73–78 (2002).
De Leenheer, E. M., Bosman, A. J., Kunst, H. P., Huygen, P. L. & Cremers, C. W. Audiological characteristics of some affected members of a Dutch DFNA13/COL11A2 family. Ann. Otol. Rhinol. Laryngol. 113, 922–929 (2004).
McGuirt, W. T., Prasad, S. D., Griffith, A. J., Kunst, H. P., Green, G. E. & Shpargel, K. B. et al. Mutations in COL11A2 cause non-syndromic hearing loss (DFNA13). Nat. Genet. 23, 413–419 (1999).
Sivakumaran, T. A., Husami, A., Kissell, D., Zhang, W., Keddache, M. & Black, A. P. et al. Performance evaluation of the next-generation sequencing approach for molecular diagnosis of hereditary hearing loss. Otolaryngol. Head Neck Surg. 148, 1007–1016 (2013).
Choi, B. Y., Park, G., Gim, J., Kim, A. R., Kim, B. J. & Kim, H. S. et al. Diagnostic application of targeted resequencing for familial nonsyndromic hearing loss. PLoS ONE 8, e68692 (2013).
Gao, X., Su, Y., Guan, L. P., Yuan, Y. Y., Huang, S. S. & Lu, Y. et al. Novel compound heterozygous TMC1 mutations associated with autosomal recessive hearing loss in a Chinese family. PLoS ONE 8, e63026 (2013).
De Keulenaer, S., Hellemans, J., Lefever, S., Renard, J. P., De Schrijver, J. & Van de Voorde, H. et al. Molecular diagnostics for congenital hearing loss including 15 deafness genes using a next generation sequencing platform. BMC Med. Genomics 5, 17 (2012).
Baek, J. I., Oh, S. K., Kim, D. B., Choi, S. Y., Kim, U. K. & Lee, K. Y. et al. Targeted massive parallel sequencing: the effective detection of novel causative mutations associated with hearing loss in small families. Orphanet. J. Rare Dis. 7, 60 (2012).
Borsani, G., DeGrandi, A., Ballabio, A., Bulfone, A., Bernard, L. & Banfi, S. et al. EYA4, a novel vertebrate gene related to Drosophila eyes absent. Hum. Mol. Genet. 8, 11–23 (1999).
Heanue, T. A., Reshef, R., Davis, R. J., Mardon, G., Oliver, G. & Tomarev, S. et al. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 13, 3231–3243 (1999).
Verstreken, M., Declau, F., Schatteman, I., Van Velzen, D., Verhoeven, K. & Van Camp, G. et al. Audiometric analysis of a Belgian family linked to the DFNA10 locus. Am. J. Otol. 21, 675–681 (2000).
Wayne, S., Robertson, N. G., DeClau, F., Chen, N., Verhoeven, K. & Prasad, S. et al. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 10, 195–200 (2001).
Hildebrand, M. S., Coman, D., Yang, T., Gardner, R. J., Rose, E. & Smith, R. J. et al. A novel splice site mutation in EYA4 causes DFNA10 hearing loss. Am. J. Med. Genet. A 143A, 1599–1604 (2007).
Pfister, M., Toth, T., Thiele, H., Haack, B., Blin, N. & Zenner, H. P. et al. A 4- bp insertion in the eya-homologous region (eyaHR) of EYA4 causes hearing impairment in a Hungarian family linked to DFNA10. Mol. Med. 8, 607–611 (2002).
Makishima, T., Madeo, A. C., Brewer, C. C., Zalewski, C. K., Butman, J. A. & Sachdev, V. et al. Nonsyndromic hearing loss DFNA10 and a novel mutation of EYA4: evidence for correlation of normal cardiac phenotype with truncating mutations of the Eya domain. Am. J. Med. Genet. A 143A, 1592–1598 (2007).
Acknowledgements
We sincerely thank all the family members for their participation and support in this study. These investigations were supported by National Natural Science Foundation of China (No. 81200749) to Sun Yi.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Sun, Y., Zhang, Z., Cheng, J. et al. A novel mutation of EYA4 in a large Chinese family with autosomal dominant middle-frequency sensorineural hearing loss by targeted exome sequencing. J Hum Genet 60, 299–304 (2015). https://doi.org/10.1038/jhg.2015.19
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.19
This article is cited by
-
Identification of a novel CNV at the EYA4 gene in a Chinese family with autosomal dominant nonsyndromic hearing loss
BMC Medical Genomics (2022)
-
A combined genome-wide association and molecular study of age-related hearing loss in H. sapiens
BMC Medicine (2021)
-
Insights into the pathophysiology of DFNA10 hearing loss associated with novel EYA4 variants
Scientific Reports (2020)
-
Prevalence and clinical features of hearing loss caused by EYA4 variants
Scientific Reports (2020)
-
Novel compound heterozygous mutations in CNGA1in a Chinese family affected with autosomal recessive retinitis pigmentosa by targeted sequencing
BMC Ophthalmology (2016)