Abstract
A screening program on a limited number of strains belonging to the Actinoallomurus genus yielded a series of new angucyclinones. NMR and MS analyses established that these compounds are characterized by an unusual lactone ring and present up to four halogens per molecule, with one congener representing the first natural product containing a trichloromethyl substitution on an aromatic system. Remarkably, this family of metabolites seems to be produced by phylogenetically distinct Actinoallomurus isolates. Because of the unique structural features and wide distribution among Actinoallomurus, we have designated these angucyclinones as allocyclinones. Allocyclinones possess interesting activity against different Gram-positive bacteria, including antibiotic-resistant strains, with antibacterial potency increasing with the number of chlorine substituents. The tetrachlorinated compound is the most abundant congener in the allocyclinone complex.
Similar content being viewed by others
Introduction
The importance of natural products to human health has been acknowledged over the past few decades and still is. Among the new chemical entities approved as drugs between 1981 and 2010, ∼68% are directly derived or inspired from natural products. This success, despite a general disinterest by pharmaceutical companies in natural products as a source of drug leads, underscores Nature’s ability to deliver complex and high-affinity chemical scaffolds that have no match in most synthetic libraries.1, 2 One therapeutic area that has been strongly dependent on natural products has been the anti-infectives, particularly antibacterial agents that have derived almost exclusively from microbial products. The past few years have seen a dramatic change in the landscape of infectious diseases, with a steady emergence and spread of antibiotic resistance giving origin to pathogens that can escape most available therapeutic options.3
The so-called golden era of antibiotics has yielded the vast majority of microbial metabolites, most of them presumably discovered because of their antibacterial potency, the amount produced by the corresponding strains and/or the natural frequency of occurrence in nature.4, 5 As these ‘low-hanging fruits’ were discovered, random screening of microbial isolates led to progressively diminished returns in terms of chemical novelty. Therefore, different approaches have been proposed and implemented to increase the probability of making metabolite discovery from microbial sources a cost-effective endeavor.6, 7, 8, 9
One possible approach to identify new metabolites is to explore microbial taxa that have not been previously systematically evaluated for the production of bioactive metabolites. In this respect, screening for antimicrobial activity may be a simple surrogate for other biological effects, as over half of the known antimicrobial compounds can exhibit other bioactivities.10 We previously reported on the use of underexplored group of actinomycetes as a promising strategy to discover novel chemistry. In particular, the genus Actinoallomurus, a recently described member of the Thermomonosporaceae within the phylum Actinobacteria, has proved to be a versatile producer of bioactive compounds, including novel ones.11, 12, 13 As part of our screening effort we came across new bioactive angucyclinones, designated allocyclinones as described herein.
Results
Identification of new angucyclinones
The procedure for screening has been previously described,11 and can be summarized by the use of strain cultivation in shake-flasks, followed by testing the resulting extracts for antimicrobial activity. Active extracts were then evaluated for the novelty of the active compound(s). With this approach, we identified Actinoallomurus sp. ID145698 as a strain worthy of further investigation. Extracts derived from this strain inhibited growth of Staphylococcus aureus and of a hyperpermeable Escherichia coli strain. After HPLC fractionation and testing of each fraction under the same conditions, LC-MS analysis indicated that the antimicrobial activity was associated with a complex of related halogenated molecules 1–4 (Supplementary Figure S1).
Structure determination
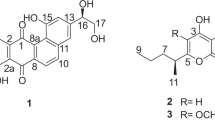
The structures of compounds 1–4 are shown in Figure 1 and were determined as follows. The UV spectra of the purified molecules were very similar with absorption up to 650 nm and the presence of three maxima at 240, 300 and 480 nm, suggesting an angucyclinone chromophore. The isotopic pattern in the electrospray ionization mass spectra clearly revealed the presence of at least one chlorine atom in all congeners. The experimental isotopic patterns and those calculated from the HR-MS-derived molecular formulas (Table 1) were in full agreement (Supplementary Figure S2) and confirmed that compounds 1 to 4 contain one to four chlorines, respectively.
1H-NMR spectra were acquired on each single, purified compound 1–4 (Supplementary Figures S3–S6). However, there was insufficient material to obtain strong carbon signals in the 2D NMR spectra that were instead obtained using the 1–4 mixture (Supplementary Figures S7–S10). A comparison of the 1H-NMR spectra of the purified compounds with the 2D NMR spectra of the mixture enabled assigning signals from the latter spectra to each congener. This approach was possible thanks to the low complexity of the proton spectra originating from a single aromatic AB system and a few singlets.
The presence of two hydrogen-bonded phenolic hydroxyls, above 12 ppm, followed by the detection of two quinone carbonyl groups at 185.6 and 187.8 ppm, confirmed the presence of a quinone. When comparing the 1H-NMR spectra of compounds 1–4, we observed a downfield shift of peaks integrating for one proton that arise from protons at positions 5 and 6 in the aromatic AB system. A similar behavior was observed for the third aromatic signal at position 4, whose chemical shifts ranged from 7.63 in 1 to 8.59 in 4, reflecting the electron withdrawing power of the substituent at the vicinal position 3, where the molecules differ (Supplementary Figure S11). Specifically, the substituents at position 3 range from CH3 in 1 to CCl3 in 4 through a stepwise replacement of each hydrogen with a chlorine atom. The corresponding protons were clearly identifiable in each 1H-NMR spectrum (Supplementary Figures S3–S6), whereas the position of the C-13 substituents was assigned through their HMBC (heteronuclear multiple bond correlation) correlations with H-4 (Supplementary Figure S9). The TOCSY (Total Correlation Spectroscopy) signals observed between CH3 at 2.66 ppm and H-4 at 7.63 ppm in compound 1, and between the chloromethylene at 4.92 ppm and H-4 at 7.92 ppm in compound 2, provided an additional confirmation (Supplementary Figure S10). On the contrary, the signals arising from common groups further away from position 3 were isochronous in compounds 1–4: two methoxy, one methyl, two phenols and a quinone moiety. The HMBC correlations between the quaternary carbon 14 and each of the methyl group at positions 17 and 15 (Supplementary Figure S9) indicated the presence of a lactone ring. The position of the additional chlorine atom present in all congeners at position 2 was also confirmed by HMBC correlations. The position of the methoxy group on ring A was suggested by a weak HMBC correlation between C-1 and H-4, consistent with a four-bond correlation reported for the related chlorocyclinones.14 This assignment was confirmed by oxidative rupture of the internal quinone with 30% H2O215, 16 that afforded a breakdown product comprising the AB ring system and the methoxy moiety (data not shown). The observed oxidation behavior is consistent with the presence of the quinone moiety in ring C. The 1H and 13C assignments are reported in Table 2.
It should be noted that the fermentation broth of strain ID145698 contained additional molecules (Supplementary Figure S1) that, on the basis of their UV spectra and MS fragmentations, appeared related to the main components 1–4. Among them, compounds 5 (Supplementary Figures S12–S14) and 6 (Supplementary Figures S15 and S16) present at position 3 a carboxylic acid and a benzylic alcohol, respectively, suggesting that they originate by formal ‘hydrolysis’ of the halogenated congeners. Indeed, 2, 3 and 4 converted into 6, 7 and 5, respectively, after several weeks at 4 °C or overnight at 60 °C (Supplementary Figure S17). Compound 7 showed m/z values consistent with the presence of an aldehyde at C-13. Furthermore, other dichlomethylene-containing compounds have been reported to convert to the corresponding aldehydes,17 suggesting that compound 7 has the structure reported in Figure 1. With hindsight, compound 7 was also detected as a minor peak in the fermentation broth of strain ID145698 (Supplementary Figure S1). Overall, these data suggest that compounds 5–7 might be just degradation products and not true pathway intermediates.
Congener distribution in Actinoallomurus sp. ID145698
Under our experimental conditions, Actinoallomurus sp. ID145698 reached stationary phase after ∼72 h, whereas angucyclinone production proceeded at a steady rate between 48 and 168 h, when 380 μg ml−1 (as the sum of compounds 1 through 4) was observed (Figure 2). Thereafter, the angucyclinone content declined (Figure 2), along with a modest increase in medium pH (data not shown). In the 72–168 h interval, the relative ratio among the four main congeners remained approximately constant, with 4 always as the main product accounting for 60–70% of the total. This observation suggests that 4 is the end product of the biosynthetic pathway and that hyperchlorination of the aromatic methyl at position 3 is not a consequence of accumulation of partially chlorinated compounds.
Time course of biomass accumulation and angucyclinone production by Actinoallomurus sp. ID145698. Columns indicate the concentrations of the individual compounds as: light blue, 1; gray, 2; yellow, 3; and dark blue, 4. The line reports biomass as percent packed mycelial volume (PMV%). Values correspond to the average of five flasks, and error bars indicate s.d. A full color version of this figure is available at The Journal of Antibiotics journal online.
Only a few chlorinated angucyclinones have been described so far: chlorocyclinones,14 marmycin B18 and JBIR-88.19 Among them, chlorocyclinone D15 is most related to the compounds described herein sharing, in addition to the chlorine at position 2 and the methoxy at position 1, a lactone fused to ring D. However, although only one member of the chlorocyclinone family contains such a lactone ring,14 this moiety is a common feature of compounds 1–4 where it is further decorated with a methoxy group. The most remarkable feature, however, is that chlorination in compounds 1–4 is not limited to the aromatic rings, as found in other chlorinated angucyclinones, but also involves the methyl group at position 3. Although aliphatic trichloromethyl groups have been previously described (for example, in the cyanobacterial product barbamide20 and in muironolide, isolated from a marine sponge21), to our knowledge 4 represents the first instance of a natural product containing an aromatic trichloromethyl substituent.
Biological activity
The entire complex (compounds 1–4) showed MIC values in the range of 0.25–1 μg ml−1 against all Gram-positive bacteria tested, with the exception of Enterococcus faecium that was approximately one order of magnitude less sensitive (Table 3). As expected, none of the evaluated, clinically relevant resistance mechanisms affected the activity of the compounds. Although there was no measurable MIC against the hyperpermeable E. coli strain used in the initial screening, growth of this strain was >50% inhibited by 32 μg ml−1 1–4 when optimal density of the culture was continuously monitored (data not shown). This observation is consistent with the activity observed on the fermentation broth extract.
The purified molecules were also evaluated for their antibacterial activities. Although only a limited number of strains were evaluated, with one exception (compounds 1 and 2 against Streptococcus pyogenes), we observed a direct relationship between the number of chlorines atoms and antibacterial activity, with each additional chlorine contributing to a 2–4-fold decrease in MIC (Table 3). Therefore, 4 is the most abundant and most active compound in the complex. In contrast, compounds 5–7 did not present noticeable antibacterial activity (data not shown).
Chlorinated angucyclinones possess different biological activities, ranging from cytotoxic as for JBIR-88 to modulators of a potential target in diabetes, like chlorocyclinones.14, 19 Further studies will be necessary to evaluate the activity of compounds 1–4 against additional biological targets.
Analysis of additional Actinoallomurus strains
We next analyzed other Actinoallomurus strains for the presence of similar compounds. Remarkably, compounds 1–4 were a relatively common encounter in Actinoallomurus, as we identified 11 additional strains (out of 200 analyzed) producing this family of angucyclinones, as indicated by similar HPLC profiles (Figure 3) and confirmed by LC-MS (Supplementary Figure S18). The 12 Actinoallomurus producers were isolated from 3 distant geographic areas and belong to 3 different phylotypes (Table 4), with 9 and 2 strains clustering with the previously described Alp24 and Alp9 phylotypes, respectively.11
HPLC traces (270 nm) of MeOH extracts from the 12 Actinoallomurus strains identified as producers of allocyclinones (Table 4). Note the 10-fold enlargement of the chromatograms for some of the extracts. A full color version of this figure is available at The Journal of Antibiotics journal online.
When analyzed in a single medium and at a single time point, all the Actinoallomurus strains produced variable amounts of the complex. However, for all strains 4 appeared to be the main congener (Figure 3), consistent with the hypothesis that this hyperchlorinated angucyclinone is the end product of the pathway. Overall, the relatively high frequency at which compounds 1–4 have been detected (that is, in 12 out of 200 screened strains) and their production by 3 phylogenetically distinct strains suggests an important role for these angucyclinones in Actinoallomurus physiology. We thus coined the name ‘allocyclinone’ for this family of compounds to reflect their frequent association with Actinoallomurus. Compound 4, the likely end product of the pathway, would then be allocyclinone A, whereas compounds 1–3 represent allocyclinones B–D, respectively (Figure 1).
Discussion
This work describes the first hyperchlorinated angucyclinone, an additional example of halogenated metabolites produced by Actinoallomurus. Indeed, this genus has been previously reported to produce spirotetronates with multiple halogens in the pyrrole moiety12 and NAI-107, a lantibiotic with a halogenated tryptophan residue.22 Although the presence of a third chlorine atom in the pyrrole moiety resulted in a lower antibacterial activity in the spirotetronate NAI-414,12 the antibacterial activity of allocyclinones increases with the number of chlorine substituents on the methyl group.
Allocyclinones might not be uncommon metabolites within the genus Actinoallomurus, as we identified several producer strains. A retrospective analysis by Baltz23 highlighted the correlation between the frequency at which different antibiotics were encountered during screening programs and the history of antibiotic discovery during the so-called ‘golden era’. It is thus not surprising that when systematically evaluating a poorly explored taxon such as Actinoallomurus, the first identified metabolites would represent the ‘low-hanging fruits’ for this genus.4 Indeed, the most abundant and most active congener, allocyclinone A, fits well within this definition: it is produced by different phylotypes (or by a frequent phylotype such as Alp24); it is produced in relatively high amounts by different isolates (>100 μg ml−1); and it has good antimicrobial activity (MIC <1 μg ml−1 against S. aureus). Compounds like this are thus hard to miss in a screening program and the low-hanging fruits within a previously unexplored taxonomic group may still provide bioactive compounds with novel structural features.
Experimental procedures
General procedures
NMR spectra were measured in CDCl3 on a Bruker (Bruker Italia Srl, Milano, Italy) 400 MHz instrument using tetramethylsilane as an internal reference. HR-MS analyses were performed on an Exactive benchtop mass spectrometer (Thermo Fisher Scientific Spa, Milano, Italy), equipped with a nanospray ionization/electrospray ionization ion source by direct infusion of compounds dissolved in MeOH–H2O 8:2. LC-MS and UV data were recorded on a Thermo LCQ equipped with heated electrospray ionization ion source with a Dionex Ultimate 3000 HPLC Column (Atlantis T3, 3 μm, 100 Å, 4.6 × 50 mm) under the following conditions: phase A, 0.1% HCOOH; phase B, MeCN; flow rate, 0.8 ml min−1; 10% phase B for 1 min followed by a linear gradient from 10 to 95% phase B in 6 min, and additional 2 min at 95% phase B. HPLC analyses were carried out using a Shimadzu LC-2010AHT (Shimadzu Corporation, Kyoto, Japan) equipped with a 5 μm (125 × 4 mm) LiChrosphere RP18 column (Merck Millipore, Darmstadt, Germany) under the following conditions: phase A, 0.1% TFA; phase B, MeCN; linear gradient from 10 to 90% phase B in 30 min; and detection at 270 nm.
Strains and growth conditions
Actinoallomurus strains were from the NAICONS collection (Table 4). Strains were maintained on S1-5.5 plates.11 For the initial screening, a loopful of mycelium was used to inoculate shake-flasks containing AF-A medium (dextrose 10 g, soybean meal 4 g, yeast extract 1 g, NaCl 0.5 g, MES 1.5 g, in 1-l deionized water, adjusted to pH 5.6 before sterilization).
For characterization of allocyclinones, frozen stocks of Actinoallomurus sp. ID145698 were used to inoculate AF-A medium and incubated for 72 h as above. Then, a 5% inoculum was transferred into fresh medium (4 × 100 ml) and incubated as above for the required amount of time. Biomass accumulation was measured as percent packed mycelial volume (PMV%) after centrifuging cultures for 10 min at 3000 r.p.m. in a graduated tube. For metabolite analysis, culture samples were mixed with 2 vol MeOH/AcOH 92:8, incubated for 1 h at room temperature and analyzed by HPLC as above after separating the residual biomass. Congener ratio was established assuming identical molar absorptions at 270 nm. Compounds concentrations were measured using pure 4 as reference standard.
PCR amplification and analysis of the 16S rRNA gene sequences were performed as previously described.24 Strains were assigned to the same phylotype when the identity of their 16S rRNA gene sequences was ⩾99.5%. Accession numbers are listed in Table 4.
Compound purification and analysis
After cultivating Actinoallomurus sp. ID145698 in medium AF-A for 192 h, the entire culture (400 ml) was acidified to pH 5 with AcOH, and centrifuged (3000 r.p.m. for 10 min) to separate the mycelium from the cleared broth. The former was treated with EtOH (100 ml) for 1 h at room temperature under shaking, then the biomass was removed by centrifugation and the supernatant was evaporated to dryness, yielding ∼100 mg of solid. The cleared broth was extracted with EtOAc (150 ml) and the organic phase was collected and dried, providing an additional 50 mg of solid. The two extracts showed similar congener distributions and were separately submitted to the following purification procedure with comparable results. Each extract was resolved by preparative TLC (Analtech Preparative Silica Gel GF with UV254 2000 μm; Sigma-Aldrich, St Louis, MO, USA) in dichloromethane–MeOH 9:1 containing 0.5% (v/v) AcOH. The main spot at Rf 0.9 was excised and compounds 1–4 were recovered by elution with MeOH, providing a total of 13 mg from both extracts. Additional spots at Rf 0.2 and 0.5 afforded 5 (4 mg) and 6 (1 mg), respectively.
An aliquot (500 μg) of the mixture 1–4 was further purified by HPLC obtaining pure 1 (65 μg), 2 (178 μg), 3 (100 μg) and 4 (176 μg). Quantities were estimated by integrating the area of the respective peak from the HPLC chromatogram.
Stability of purified 2, 3 and 4 was assessed at 10 mg ml−1 in DMSO. Solutions were analyzed by LC-MS before and after heating overnight at 60 °C.
Antimicrobial assays
All MICs were determined by broth microdilution in sterile 96-well microtiter plates, using Müller Hinton broth (Difco Laboratories, Becton, Dickinson and Company, Franklin Lakes, NJ, USA) containing 20 mg l−1 CaCl2 and 10 mg l−1 MgCl2 for all strains except for Streptococcus spp. that were grown in Todd Hewitt broth (Difco Laboratories). Strains were inoculated at 5 × 105 CFUs per ml and incubated at 37 °C for 20 h. For growth inhibition of E. coli L4242 (a ΔtolC derivative of MG1641), the same procedure was used except the OD590 was continuously measured in a BioTek Synergy2 plate reader (BioTek Instruments, Winooski, VT, USA). Compounds were dissolved in DMSO at 10 mg ml−1 and diluted with the culture medium immediately before testing. All strains were either from the ATCC (Manassas, VA, USA) or the NAICONS collection and relevant resistance phenotypes are reported in Table 3.
References
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 (2012).
Ganesan, A. The impact of natural products upon modern drug discovery. Curr. Opin. Chem. Biol. 12, 306–317 (2008).
Wright, G. D. Antibiotics: a new hope. Chem. Biol. 19, 3–10 (2012).
Monciardini, P., Iorio, M., Maffioli, S., Sosio, M. & Donadio, S. Discovering new bioactive molecules from microbial sources. Microb. Biotechnol. 7, 209–220 (2014).
Baltz, R. H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8, 557–563 (2008).
Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015).
Pena, I. et al. New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: an open resource. Sci. Rep. 5, 8771 (2015).
Donadio, S., Monciardini, P. & Sosio, M. Chapter 1. Approaches to discovering novel antibacterial and antifungal agents. Methods Enzymol. 458, 3–28 (2009).
Gengenbacher, M. & Dick, T. Antibacterial drug discovery: doing it right. Chem. Biol. 22, 5–6 (2015).
Berdy, J. Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. 65, 385–395 (2012).
Pozzi, R. et al. The genus Actinoallomurus and some of its metabolites. J. Antibiot. 64, 133–139 (2011).
Mazzetti, C. et al. Halogenated spirotetronates from Actinoallomurus. J. Nat. Prod. 75, 1044–1050.
Inahashi, Y. et al. Actinoallolides A-E, new anti-trypanosomal macrolides, produced by an endophytic actinomycete, Actinoallomurus fulvus MK10-036. Org. Lett. 17, 864–867 (2015).
Potterat, O. et al. Chlorocyclinones A-D, chlorinated angucyclinones from Streptomyces sp. strongly antagonizing rosiglitazone-induced PPAR-gamma activation. J. Nat. Prod. 70, 1934–1938 (2007).
Thomson, R. H. Naturally Occurring Quinones, (Academic Press, (1971).
Ogawa, H. & Natori, S. Hydroxybenzoquinones from Myrsinaceae plants. III. The structures of 2-hydroxy-5-methoxy-3-pentadecenylbenzoquinone and ardisiquinones A, B, and C from Ardisia spp. Chem. Pharm. Bull. 16, 1709–1720 (1968).
Li, W., Li, J., DeVicentis, D. & Mansour, T.S. Oxygen transfer from sulfoxide: formation of aromatic aldehydes from dihalomethylarenes. Tetrahedron Lett. 45, 1071–1074 (2004).
Martin, G. D. et al. Marmycins A and B, cytotoxic pentacyclic C-glycosides from a marine sediment-derived actinomycete related to the genus Streptomyces. J. Nat. Prod. 70, 1406–1409 (2007).
Motohashi, K., Takagi, M., Yamamura, H., Hayakawa, M. & Shin-ya, K. A new angucycline and a new butenolide isolated from lichen-derived Streptomyces spp. J. Antibiot. 63, 545–548 (2010).
Gerwick, W. H. & Orjala, J. Barbamide, a chlorinated metabolite with molluscicidal activity from the Caribbean cyanobacterium Lyngbya majuscula. J. Nat. Prod. 59, 427–430 (1996).
Dalisay, S. D., Morinaka, B. I., Skepper, C. K. & Molinski, T. F. A tetrachloro polyketide hexahydro-1H-isoindolone, muironolide A, from the marine sponge Phorbas sp. natural products at the nanomole scale. J. Am. Chem. Soc. 131, 7552–7553 (2009).
Cruz, J.C.S. et al. Brominated variant of the lantibiotic NAI-107 with enhanced antibacterial potency. J. Nat. Prod. 78, 2642–2647 (2015).
Baltz, R. H. Antimicrobials from actinomycetes: back to the future. ASM Microbe 2, 125–131 (2007).
Monciardini, P., Sosio, M., Cavaletti, L., Chiocchini, C. & Donadio, S. New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes. FEMS Microbiol. Ecol. 42, 419–429 (2002).
Acknowledgements
The research leading to these results was partially supported by the European Community’s Seventh Framework Programme (FP7/2007-2013) under Grant Agreement No. 289285. JCSC was entirely supported by this Grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest in addition to their affiliations to KtedoGen and/or NAICONS.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Cruz, J., Maffioli, S., Bernasconi, A. et al. Allocyclinones, hyperchlorinated angucyclinones from Actinoallomurus. J Antibiot 70, 73–78 (2017). https://doi.org/10.1038/ja.2016.62
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2016.62
This article is cited by
-
Promising bioactive compounds from the marine environment and their potential effects on various diseases
Journal of Genetic Engineering and Biotechnology (2022)