Abstract
Despite that three types of assays (measuring biofilm viability, biomass, or matrix) are described to assess anti-biofilm activity, they are rarely used together. As infections can easily reappear if the matrix is not affected after antibiotic treatments, our goal was to explore the simultaneous effects of antibiotics on the viability, biomass and matrix of Staphylococcus aureus biofilms (ATCC 25923). Viability and biomass were quantified using resazurin and crystal violet staining sequentially in the same plate, while matrix staining was conducted with a wheat germ agglutinin-Alexa Fluor 488 fluorescent conjugate. Establishment of the detection limits and linearity ranges allowed concluding that all three methods were able to estimate biofilm formation in a similar fashion. In a susceptibility study with 18-h biofilms, two model compounds (penicillin G and ciprofloxacin) caused a reduction on the viability and biomass accompanied by an increase or not changed levels of the matrix, respectively. This response pattern was also proven for S. aureus Newman, S. epidermidis and E. coli biofilms. A classification of antibiotics based on five categories according to their effects on viability and matrix has been proposed earlier. Our data suggests a sixth group, represented by penicillin, causing decrease in bacterial viability but showing stimulatory effects on the matrix. Further, if effects on the matrix are not taken into account, the long-term chemotherapeutic effect of antibiotics can be jeopardized in spite of the positive effects on biofilms viability and biomass. Thus, measuring all these three endpoints simultaneously provide a more complete and accurate picture.
Similar content being viewed by others
Introduction
Bacterial biofilms are sessile cellular communities attached to solid surfaces and embedded in a self-produced matrix of extracellular polysaccharides (EPS). The matrix is important, as it provides structural stability and protection to the biofilm. It acts as a protective barrier against biocides or toxins and it sequesters nutrients from the environment, so therefore it is part of the general bacterial strategy for persistence under extreme, non-favorable conditions.1 The matrix composition is known to differ between and within species, but it mainly consists of polysaccharides, DNA and proteins.2 The biofilms exhibit increased resistance towards antimicrobial therapy when compared with their suspension counterparts. Thus, finding anti-biofilm molecules has proven to be a tremendous challenge. In fact, a limited repertoire of antimicrobials exists today able to kill mature biofilms, particularly in the case of Staphylococcus spp.3
The success of finding effective drugs for any given target greatly depends on the chosen bioassays. For biofilms, three types of surrogate whole-cell assays in 96-well plates have been described based on quantification of biomass as well as biofilm components: viable cells and EPS. For characterizing anti-biofilms activity, the most common practice has been to use a single type of assay that measures reduction of total biomass or cellular viability4, 5 with less emphasis put into combining the three assays.6 One likely explanation is that, to measure the EPS content, most of the available methods depend on the extraction of polysaccharides,7, 8 which makes them more laborious and time-consuming for anti-biofilm susceptibility studies.
Typically available antibiotics act on metabolically active bacteria that are not commonly found in the EPS. Thus, antibiotics can kill biofilm cells but cause other effects on the EPS fraction. It has been suggested that surfaces in which the matrix has been left, can be recolonized more rapidly by biofilm-forming bacteria,9, 10, 11 causing infection reappearance. In two previous contributions it has been demonstrated that some antibiotics (that is, vancomycin), at millimolar concentrations, decrease the bacterial viability without having a significant effect on the matrix.10, 11 In such studies, along with the conventional resazurin assay, a method to estimate the matrix content was used, based on the staining with dimethyl methylene blue, a cationic dye that stains the polysaccharide intercellular adhesin in Gram-positive biofilms.9
Given the tremendous implications of those mentioned studies for the critical assessment of the long chemotherapeutical effects of conventional antibiotics, we decided, in the present contribution, to further explore the effects of antibiotics on the matrix, viability and biomass of S. aureus biofilms. First, to exclude the possibility that previous results could have been specifically related to the use of dimethyl methylene blue (which could for instance interact with the antibiotics, leading to overestimation of the matrix effects), we replaced this assay with another type of method originally described by Burton et al.,12 that has the advantage of being applicable to both Gram-positive and Gram-negative bacteria without involving polysaccharide extraction. The assay utilizes a wheat germ agglutinin-Alexa Fluor 488 fluorescent conjugate (WGA) that binds to poly-N-acetylglucosamine (PNAG) residues present in the typical EPS of Staphylococcus spp. biofilms. This conjugate binds specifically to N-acetylneuraminic acid in the peptidoglycan layer of Gram-positive bacteria as well as to polysaccharide adhesin involved in biofilm formation by both Gram-positive and -negative bacteria.12
Second, we applied this WGA assay, along with crystal violet and resazurin stainings to have an overall picture of the changes taking place in the matrix, biomass and viability, respectively, of Staphylococcus aureus biofilms when exposed to two other model antibiotics: penicillin and ciprofloxacin. Bearing in mind that removal of the matrix is as important as killing of the bacteria to ensure good clinical anti-biofilm effects, our goal here was to establish if contradictory results could be obtained between the three (not only two) assays that could jeopardize the efficacy of antibiotics in biofilms infections.
In third place, we choose another biofilm-forming Staphylococcus aureus strain different to the ones included in previous studies and confirmed the importance of PNAG in the matrix of these biofilms. We also decided to utilize a wider concentration range for the model antibiotics, covering from sub-inhibitory concentrations (500 pM) to millimolar levels (5 mM), so that the whole spectrum of possible actions, including the clinical ones, could be covered and comparable conclusions could be drawn on the basic actions of commonly used antibiotics.
Materials and methods
Bacterial growth and biofilm formation
The biofilm-producing strains of Staphylococcus aureus (ATCC 25923 and Newman, both clinical strains), Staphylococcus epidermidis (ATCC 12228, non-clinical and ATCC 35984, clinical strain) and Escherichia coli (XL1 Blue, non-clinical) were provided by the Faculty of Pharmacy or Faculty of Veterinary Medicine, University of Helsinki, Finland. Bacteria were cultured in tryptic soy broth (Fluka Biochemika, Buchs, Switzerland) under aerobic conditions at 37 °C and 200 r.p.m. to reach exponential growth, typically up to a concentration of 108 CFU ml−1. This was estimated by spectrophotometric turbidity measurement at 595 nm using a Varioskan Multimode Plate Reader (Thermo Fisher Scientific Oy, Vantaa, Finland) and by CFU counts on tryptic soy agar (Fluka Biochemika) plates. Biofilms of all strains were formed by addition of 106 CFU ml−1 of the bacterial suspension in sterile, flat-bottomed, 96-well polystyrene micro-well plates (Nunclon Δ surface; Nunc, Roskilde, Denmark) (200 μl per well) and incubated for 18 h under aerobic conditions (37 °C, 200 r.p.m. shaking).
Platform of three biofilm quantification assays
For measuring the presence of viable cells and the total biomass in the wells, resazurin staining and crystal violet were used, respectively, as described earlier.13, 14 Briefly, the biofilms were first stained with 20 mM resazurin for 20 min in room temperature (RT), 200 r.p.m. and fluorescence was measured at λexcitation=560 nm and λemission=590 nm, when using the S. aureus and S. epidermidis strains. When using E. coli, the staining time was prolonged to 35 min. Then, the resazurin stain was removed and replaced by crystal violet stain that was added in the same plate. The biofilms were stained for 5 min (RT) and then washed three times with Milli-Q-water using the automated protocol.13 The remaining dye was then solubilized in 96% ethanol and absorbance at 595 nm was measured. The WGA method was conducted in a replicate plate as originally described12 with modifications. Before adding the stain, the wells were washed once with sterile PBS, then 200 μl of 5 μg ml−1 WGA (WGA-Alexa Fluor 488 fluorescent conjugate, Molecular Probes Inc., Eugene, OR, USA) in PBS was added and the biofilms were incubated for 2 h at 4 °C in the darkness. After removing the unbound dye, plates were washed three times with PBS, air-dried for 15 min (RT) and the bound probe was solubilized with 200 μl of 33% acetic acid. Plates were covered with strip caps and sonicated two times for 30 s with 1 h incubation period at 37 °C in between. Then, fluorescence at λexcitation=495 nm and λemission=520 nm was measured with a Varioskan Multimode Plate Reader.
Establishment of the detection limits of the assays
The relation between the actual biofilm concentrations on the wells and the endpoint signal generated by each assay was first measured. For that, different biofilm concentrations were achieved by incubating bacterial suspensions (106 CFU ml−1) on flat-bottomed 96-well polystyrene micro-well plates at 37 °C, 200 r.p.m. during different times. The plates were then endpoint stained using the three different methods (resazurin, crystal violet and WGA assays), as described above. To determine the actual concentrations using the conventional agar plate counts (CFU ml−1), biofilms from replicate wells from each assay were vigorously scraped, serially diluted in tryptic soy broth and plated on tryptic soy agar.
Susceptibility trials
The effect of three model compounds (penicillin G, ciprofloxacin and rosmarinic acid) displaying known effects on S. aureus biofilms, was assessed. Bacterial suspensions (106 CFU ml−1, 200 μl per well) were added to the plates and maintained at 37 °C, 200 r.p.m., during 18 h in tryptic soy broth to form biofilms. Compounds (500 pM–5 mM) were added at the same time with the bacterial suspensions (preventive effect) or added onto mature biofilms, previously formed for 18 h in tryptic soy broth (destruction effect). Mature biofilms were incubated with the compounds at 37 °C, 200 r.p.m. for further 24 h. At the end of the exposures, biofilms were stained with resazurin and crystal violet (in the same plate) and the WGA probe (in a replicate plate), as indicated before. In some experiments, mature biofilms (18 h) were treated with 2 μg ml−1 of proteinase K (in 50 mM Tris buffer, 1 mM EDTA) for 6 h and 22 h at 37 °C or with 0.02 U ml−1 of DNase I (in 40 mM Tris buffer, 1 mM CaCl2, 10 mM MgCl2) for 24 h at 37 °C, to determine the role of extracellular proteins or extracellular DNA (eDNA), respectively, in the matrix. To exclude the possibility that the antibiotics could have some unspecific interactions with the WGA probe, the compounds were incubated with the WGA for 2 h at 4 °C and then fluorescence was measured at λexcitation=495 nm and λemission=520 nm.
Visualization of the effect on the biofilms using fluorescence microscopy
Mature (18-h) biofilms exposed to penicillin (400 μM) were imaged for viability and EPS production using fluorescence microscopy. Viability imaging was done with the commercial bacterial viability kit LIVE/DEAD BacLight (Molecular Probes Inc.) that contains SYTO 9 (stains viable cells green) and propidium iodide (stains dead cells red). Final concentrations of the probes (added as a mixture, 6 μl per well) were 5 μM and 30 μM, respectively. For EPS imaging, the WGA probe was added and incubated for 2 h at 4 °C in darkness, as previously described. Fluorescence microscopy pictures were taken after removing the unbound dye. Images were captured with an AxioVert 200 M fluorescence microscope (Carl Zeiss MicroImaging GmbH, Munich, Germany), using a FITC filter (Syto 9, WGA) or a TRITC filter (propidium iodide).
Statistical analysis
All exposure experiments were done with at least three biological replicates in triplicates. The results are shown as per cent of control±s.d. Assay performance was measured using typical statistical parameters: screening window coefficient Z′-factor, signal-to-noise (S/N) and signal-to-background (S/B) (Equations 1, 2, 3). One-way analysis of variance comparisons and Tukey post tests were applied to the original fluorescence and absorbance data points using GraphPad Prism software, v. 5.0 for Mac OS (La Jolla, CA, USA). In the case of paired comparisons, unpaired t test with Welch's correction was used. P<0.05 was considered statistically significant. Coefficient of variations (CV) of the signals was calculated using Equation 4.




Results and discussion
We have previously described two staining assays for measuring total biomass and viability of biofilms using crystal violet and resazurin, respectively,13, 14 and proposed the use of these two combined assays as a first-tier bioactivity profiling platform to measure anti-biofilm effects in a screening-compatible setting. In this contribution, we introduced a new practical improvement to the platform by performing both assays in sequential workflow using the same plate, instead of running the assays in separate plates.
Staining the biofilms in the same plate first with resazurin and then with crystal violet, reduces the usage of tested compounds, reagents (that is, culture media) and consumables (that is, tips, plates) significantly and the time required for each experiment. Resazurin is known to be a non-invasive dye that leaves cells intact,15 and indeed no effect of the resazurin staining could be detected in the amount of biomass stained by crystal violet assay when compared with the staining performed in separate plates. The maximal signal achieved for biofilms stained with crystal violet performed in separated plates was 1.57±0.29 (relative absorbance units, RAU), whereas the signal measured for the crystal violet staining performed in the same plate stained with resazurin was 1.52±0.25 (RAU). Thus, no statistically significant changes were detected in the amount of stained biomass (P=0.4149).
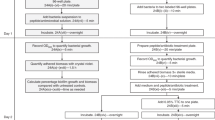
By combining resazurin and crystal violet assays, more relevant information on the mechanisms of the antibiotics can be obtained than when using the separate assays. For instance, compounds with general bactericidal actions, tend to cause a decrease in the resazurin reduction and the crystal violet stained biomass of exposed biofilms, but compounds with detergent-like actions (and no bactericidal effects), tend to decrease the biomass without causing effects on the alive cells in the core of the biofilms. These types of compounds could easily be distinguished by applying these two methods. In addition, both assays can be applied to the rapid screening of compounds in prevention of biofilm formation and destruction of mature biofilms, as they are not expensive, or too laborious. However, discrimination of specific actions of antibiotics on the biofilm matrix would still not be possible. Thus, we added as a third method, the WGA assay, to estimate the EPS content. The combination of the three assays can then ensure that a broader view on anti-biofilms effects is achieved. For a practical viewpoint, we recommend the resazurin and crystal violet assays to be done sequentially in the same plate, while a parallel set of samples can be tested with the WGA assay. This workflow is schematically presented in Figure 1.
Schematic representation of the workflow proposed for the characterization of anti-biofilm compounds in micro-well plates. NA and A stand for treatment with non-active compounds and active compounds, respectively. Active compound is defined in the figure as a hit compound able to reduce the viability, biomass and EPS of the biofilms. The opposite applies to a non-active compound. Fluorescence-based endpoint measurements in the case of resazurin and WGA assays are indicated with yellow stars. A full color version of this figure is available at The Journal of Antibiotics journal online.
A common reason for divergences between experimental assays characterizing the same biological process is that they may differ in their detection limits. Therefore, to make sure here that the three assays could consistently detect biofilm formation on the micro plate wells within similar bacterial concentration ranges, signal generated in each case was compared with actual bacterial counts obtained from scratched wells (Figure 2). Given their different performances when using different strain types or microorganisms, attention was paid to their detection limits as well as linearity ranges. The WGA assay produced a significant fluorescent signal when S. aureus biofilms were higher than 2 × 107 CFU ml−1 in the wells, which coincided with the detection limit of the previously described resazurin assay as well.16, 17 The crystal violet assay has been extensively used,17, 18, 19 but likely because the estimated biomass does not distinguish between living and dead cells, the detection limit has not been emphasized on. All three assays were able to detect biofilm formation at the typical cell densities (>107 CFU cm−2) achieved in micro-well plates assays.20 In routine biofilm susceptibility trials, when optimal biofilm formation takes place (that is, after 18 h in our assay conditions), the registered signals generated by the three assays corresponded roughly to 4–7 × 109 CFU ml−1 (biofilms) in the wells. The linearity phase of the three methods extended to fairly high S. aureus biofilm concentrations, close to 1010 CFU ml−1, thus antimicrobial efficacy of at least 3-log units is measurable with the three indicated endpoints. Performance wise, robust cell-based assays were obtained in the three cases (Z′⩾0.47; S/N⩾6.20 and S/B⩾10.60), with a quality that met the expectations for whole-cell screening methods.
Assessment of the actual biofilm concentration corresponding to the endpoint signal generated by staining the biomass with the crystal violet assay (a), measuring the viability of the biofilms with resazurin assay (b) and determining the EPS layer with the WGA assay (c). RFU stands for relative fluorescence units and RAU for relative absorbance units.
In attempting to combine these three methods for susceptibility studies, their limitations should also be acknowledged. The drawbacks of the resazurin and crystal violet assay have been discussed by us and others.16, 17 For the resazurin staining, one of the most important limitations is the need for optimization of the staining conditions (particularly the exposure time to the dye) for different bacterial strains and species. Directly applying one resazurin staining protocol from one species to another is dramatically hampered by the different rates in which this probe can be metabolized in different bacteria. Here, for instance, the staining time had to be prolonged from 20 min to 35 min when using E. coli biofilms compared with Staphylococcus spp. On the other hand, the main drawbacks of the crystal violet assay are given by the facts that only biomass can be measured, which is less informative than viability, along with the laborious performance of the method that involves several washing steps. An attempt to circumvent this problem has been the automation of the crystal violet protocol with the aid of a liquid handling robot, which was earlier reported by our group.13
In the case of the WGA assay, a likely limitation is that it may underestimate the amount of EPS in strains in which the PNAG is not the major component of the EPS. The model strain used here (S. aureus ATCC 25923) apparently forms biofilms that are mainly composed of the PNAG. In this strain, the eDNA does not seem to be a critical constituent of the EPS, as the exposure of 18-h formed biofilms to DNase I (0.02 U ml−1 for 24 h) caused no effect on the viability of mature biofilms (0% of inhibition). The model strain did not seem to be critically dependent on the presence of proteins in the EPS either, as treatment of 18-h biofilms with proteinase K (2 μg ml−1) caused only a 23.9±9.1% inhibition of bacterial viability after 6 h treatment and 13.7±8.4% of inhibition after 22 h. Situation can obviously be different in other biofilm-forming species or strains, as it has been widely supported that the biofilm matrix differ in composition and can contain varying amount of polysaccharides, eDNA and proteins in the matrix. In addition, their functional roles in the EPS layer can differ.21 Thus, as for the dimethyl methylene blue method, the application of the WGA assay depends on the presence of PNAG in mature biofilms of the bacterial strains and the interpretation of the data should also take into account the importance of the PNAG component in the matrix.
Subsequently, with these three assays, a typical susceptibility study involving three model compounds: penicillin and ciprofloxacin as active antibiotics and the weak antibiotic substance rosmarinic acid as negative control was conducted. When testing the ability to prevent biofilm formation, rosmarinic acid caused no significant changes in biomass or EPS and only a weak (not statistically significant) reduction of the viability. In contrast, the conventional antibiotics penicillin G and ciprofloxacin significantly reduced the overall biofilm biomass and the amount of viable cells, together with the EPS context, as measured by WGA staining (Figure 3a). In these conditions, it is expected that killing of the suspended cells by penicillin G and ciprofloxacin occurs mostly before the biofilms are formed. Thus, the lower the amount of suspended cells that are left alive, the lower the amount of viable cells left to form biofilms will be, as well as the biomass and EPS that they will produce.
Effect of two model compounds (penicillin G and ciprofloxacin) in (a) prevention of and (b) destruction of S. aureus biofilms viability, biomass and EPS production, measured using resazurin (black bars), crystal violet photometric staining (gray bars) and WGA fluorescent assays (white bars), respectively. Percentages of biofilm formation in relation to bacterial controls (not exposed to antimicrobials) are presented here. The left cluster corresponds to penicillin-treated biofilms and the rightmost cluster to ciprofloxacin-treated biofilms. s.d. are not shown in the figure but calcd coefficient of variation did not exceed 30%. One-way analysis of variance comparisons and Tukey post tests were applied to the original data points but for better clarification, only the results of the comparisons between the untreated biofilms and the highest concentration of the two compounds are presented here. ***A high statistically significant difference from the control, P<0.001.
However, as antibiotics are mostly administered in clinical practice to counteract an existing infection, evaluating their in vitro effects on existing biofilms provides more realistic information (Figure 3b). In these new conditions, no protective effect was registered after exposure to rosmarinic acid, in agreement with its weak antimicrobial activity.22 On the other hand, penicillin G and ciprofloxacin caused a statistically significant reduction of the biomass and cell viability of mature biofilms, but these effects were associated to a significant increase (in case of penicillin) or maintained levels (for ciprofloxacin) of the PNAG content of the EPS (Figure 3b). Addition of penicillin, for instance, at concentrations ranging from 0.5 μM to 5 mM, caused a statistically significant reduction of the biomass and viability to a plateau value around 50%. However, the EPS values, as measured by the WGA signal, rose significantly to values over 200%. Further on, the pattern of microbial response to penicillin was also confirmed using fluorescence microscopy (Figure 4). Staining with LIVE/DEAD BacLight kit (Figure 4a) corroborated that penicillin treatment (400 μM) results in nearly 50% reduction of viable cells in comparison to untreated biofilms. In contrast, the cells remaining alive produced a significantly higher amount of EPS as judged from the detected increase in green fluorescence, when compared with untreated biofilms (Figure 4b).
Fluorescence microscopy pictures of the effect of penicillin addition (400 μM) on viability and EPS production are shown here, top images (a) correspond to untreated biofilms (left) and penicillin-treated cells (right) stained with the LIVE/DEAD BacLight kit with Syto 9 for staining viable cells and propidium iodide for dead cells. The bottom images (b) correspond to untreated biofilms (left) and penicillin-treated cells (right) stained with the WGA probe. The scale bar is equal to 100 μm in all images. A full color version of this figure is available at The Journal of Antibiotics journal online.
Given the fact that only the WGA assay gave a divergent result when compared with the crystal violet and resazurin methods in 18-h biofilms treated with penicillin or ciprofloxacin, it was first questioned whether the increased WGA signal could be given by an unspecific interaction of the antibiotics with the fluorescent probe. However, the incubation of the WGA with the three antibiotics at the same four concentrations tested here, caused no changes on the intrinsic fluorescence of the WGA (results not shown), allowing to exclude that it could cause an experimental artifact.
Second, it could be argued that the detected increase of the WGA signal reflected only the increase of the PNAG content in biofilms treated with penicillin, which could be potentially accompanied by a similar or even higher decrease in the other components of the biofilm matrix (eDNA and proteins), thus ensuring that the overall levels of the EPS are decreased or at least maintained, when compared with untreated bacterial controls. However, given the fact that PNAG seems to be the predominant component of the EPS in the model strain used here and the changes in the PNAG levels were significantly high, chances that a compensatory mechanism would altogether decrease EPS content, seem fairly low to us. In fact, after extensive literature search, no studies supporting the decrease on the eDNA or protein levels of the EPS on antibiotics-treated biofilms have been found.
A third argument that could be taken up for discussion is the fact that WGA could potentially stain the EPS from dead cells. However, biofilms seem to be killed by both penicillin and ciprofloxacin (as supported by the results with the resazurin assay presented in Figure 3) and in the case of the destruction assay, there is even less amount of viable cells in ciprofloxacin-treated biofilms (30.8% of viable cells), as opposed to penicillin-treated biofilms (54.7% of viable cells). Thus, the amount of dead cells is higher in ciprofloxacin-treated biofilms, but yet the detected WGA signal is increased only after treatment with penicillin, not ciprofloxacin. This points out to the fact that WGA signal is not probably connected to the amount of dead cells.
If the possibility that the EPS levels indeed increase upon biofilms exposure to certain antibiotics is then accepted as the most likely scenario, it could be additionally claimed that this is only a strain-specific finding, not observable in other Staphylococcal strains or other bacterial species. Thus, we expanded the study to include three biofilm-forming Staphylococcus spp. strains: the S. aureus Newman and two S. epidermidis strains (ATCC 12228 and RP62A). Additionally, as a Gram-negative model strain E. coli (XL1 Blue) was also tested (Table 1). The production of the PNAG, which mediates many functions of biofilm formation, is regulated by the icaADBC operon in staphylococci23 and by the pgaABCD operon in E. coli.24 The S. aureus Newman, S. epidermidis RP62A and the E. coli XL 1 Blue strains are producers of PNAG and the WGA assay is well applicable to them.25, 26, 27 The S. epidermidis 12228 is negative for the ica locus and it is often referred to as a non-biofilm or weak biofilm producer.27, 28 In the absence of PNAG, the WGA probe is probably unable to bind to the matrix of the biofilms formed by the S. epidermidis 12228 and when using this strain, highly diverting signals, including negative Z′-values were in fact registered, thus impeding the application of the WGA assay. This strain or other non-PNAG-producer strains should therefore be carefully evaluated before applying the WGA method.
On the other hand, the performance of the WGA assay using the other strains did not differ from the model strain. The mean of the Z′-values obtained was 0.47±0.13. For the S. aureus Newman and the E. coli XL1 Blue strains (Table 1), a significant increase on EPS production, as measured by WGA signal, was registered upon the exposure of 18-h biofilms to penicillin treatment, similar to the response observed with the model S. aureus strain. Thus, from these results it could be concluded that the increases in the WGA signals observed for the model S. aureus strain were not an exclusive situation unique to that organism. In spite of the differences between the resazurin and the crystal violet assays, a similar trend of the results from the compound treatments could be seen (Table 1). If the viability of the biofilms decreased also the biomass was reduced, although it was generally observed that the viability decreased more. In the case of ciprofloxacin-treated S. aureus Newman for instance, the resazurin signal was much lower (6.4±1.2; % of control) than the crystal violet signal (63.1±6.8; % of control), which could be explained by the fact that resazurin only measures the living bacteria but the dead cells in the biomass stained with crystal violet are not taken into account.20 An exception to this could be seen for penicillin-treated S. epidermidis RP62A, where the resazurin was higher (78.0±5.1, % of control) than the crystal violet (52.3±3.3, % of control), but the trend of decreasing in both cases was still preserved.
The preservation of EPS is a postulated resource of bacterial cells in biofilms to account for their long-term survival in harsh conditions.9, 10, 11 The fact that the increase or maintained levels of the EPS content could take place after exposure to penicillin G and ciprofloxacin, which display two completely different mechanisms of actions on S. aureus biofilms, inclines us to hypothesize that this may be a more general protective response against chemical insults. Our results also support the previous views proposed by Toté et al.10 using a completely different method of EPS staining and other bacterial strains. These authors proposed a classification of antibiotics based on five categories according to their effects on matrix and bacterial viability. In category A, they included antibiotics displaying no effects on either endpoint, whereas categories B and C were dedicated to compounds with exclusive effects on the matrix (reduction) or the bacterial viability (reduction), respectively. Category D included antibiotics able to reduce both the viability and the matrix. Finally, the last category (E) was assigned to antibiotics reducing the matrix but causing other effects on the viability: at high concentrations such compounds have an inhibitory activity on viability but at sub-inhibitory concentrations they display a stimulatory effect. One of our model antibiotics, ciprofloxacin, was proven to fit into category C described by Toté et al.10 (significant effects on viability but no effects on matrix). On the other hand, our data allows to tentatively suggest the inclusion of a sixth group (which could be regarded as category F), represented by penicillin, in which significant inhibitory effects could be detected on the bacterial viability accompanied by significant stimulatory effects on matrix production.
In terms of antimicrobial characterization, our results implicate that compounds that may be regarded effective only on the basis of their positive effects on biomass and/or cell viability, could promote, on the other hand, overproduction or simply maintenance of the biofilm matrix, thus bearing a detrimental long-term chemotherapeutic effect. We have proven here that a matrix detection assay, such as the WGA method, could unravel these important elements of the biofilms response towards antibiotics that are critical to quickly assess the clinical relevance of chemotherapy against S. aureus biofilms. Such elements can be overlooked when measuring only biomass and/or viability, and therefore the addition of the WGA assay, or any other matrix detection method, should be regarded as critical during the study of anti-biofilm molecules.
References
Donlan, R. & Costerton, J. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 (2002).
Steinberger, R. E. & Holden, P. A. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 71, 5404–5410 (2005).
Landini, P., Antoniani, D., Burgess, J. & Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 86, 813–823 (2010).
Kiran, M. et al. Discovery of a quorum-sensing inhibitor of drug-resistant staphylococcal infections by structure-based virtual screening. Mol. Pharmacol. 73, 1578–1586 (2008).
Opperman, T. J. et al. Aryl rhodanines specifically inhibit staphylococcal and enterococcal biofilm formation. Antimicrob. Agents Chemother. 53, 4357–4367 (2009).
Zhao, T. & Liu, Y. N-acetylcysteine inhibit biofilms produced by Pseudomonas aeruginosa. BMC Microbiol. 10, 140 (2010).
Mack, D. et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178, 175–183 (1996).
Sadovskaya, I., Vinogradov, E., Flahaut, S., Kogan, G. & Jabbouri, S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect. Immun. 73, 3007–3017 (2005).
Toté, K., Vanden Berghe, D., Maes, L. & Cos, P. A new colorimetric microtitre model for the detection of Staphylococcus aureus biofilms. Lett. Appl. Microbiol. 46, 249–254 (2008).
Toté, K. et al. Inhibitory efficacy of various antibiotics on matrix and viable mass of Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Int. J. Antimicrob. Agents 33, 525–531 (2009).
Toté, K., Horemans, T., Vanden B., D., Maes, L. & Cos, P. Inhibitory effect of biocides on the viable masses and matrices of Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 76, 3135–3142 (2010).
Burton, E., Yakandawala, N., LoVetri, K. & Madhyastha, M. A microplate spectrofluorometric assay for bacterial biofilms. J. Ind. Microbiol. Biotechnol. 34, 1–4 (2007).
Sandberg, M., Määttänen, A., Peltonen, J., Vuorela, P. & Fallarero, A. Automating a 96-well microtitre plate model for Staphylococcus aureus biofilms: an approach to screening of natural antimicrobial compounds. Int. J. Antimicrob. Agents 32, 233–240 (2008).
Sandberg, M. E. et al. Pros and cons of using resazurin staining for quantification of viable Staphylococcus aureus biofilms in a screening assay. J. Microbiol. Methods 78, 104–106 (2009).
O'Brien, J., Wilson, I., Orton, T. & Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cytotoxicity. Eur. J. Biochem. 267, 5421–5426 (2000).
Mariscal, A., Lopez-Gigosos, R., Carnero-Varo, M. & Fernandez-Crehuet, J. Fluorescent assay based on resazurin for detection of activity of disinfectants against bacterial biofilm. Appl. Microbiol. Biotechnol. 82, 773–783 (2009).
Peeters, E., Nelis, H. & Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 72, 157–165 (2008).
Christensen, G. D. et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22, 996–1006 (1985).
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B. & Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40, 175–179 (2000).
Pitts, B., Hamilton, M., Zelver, N. & Stewart, P. A microtiter-plate screening method for biofilm disinfection and removal. J. Microbiol. Methods 54, 269–276 (2003).
Steinberger, R. E. & Holden, P. A. Macromolecular composition of unsaturated Pseudomonas aeruginosa biofilms with time and carbon source. Biofilms 1, 37–47 (2004).
Mencherini, T., Picerno, P., Scesa, C. & Aquino, R. Triterpene, antioxidant, and antimicrobial compounds from Melissa officinalis. J. Nat. Prod. 70, 1889–1894 (2007).
Gerke, C., Kraft, A., Süssmuth, R., Schweitzer, O. & Götz, F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273, 18586–18593 (1998).
Cerca, N. & Jefferson, K. K. Effect of growth conditions on poly-N-acetylglucosamine expression and biofilm formation in Escherichia coli. FEMS Microbiol. Lett. 283, 36–41 (2008).
Kropec, A. et al. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect. Immun. 73, 6868–6876 (2005).
Zmantar, T., Kouidhi, B., Miladi, H., Mahdouani, K. & Bakhrouf, A. A microtiter plate assay for Staphylococcus aureus biofilm quantification at various pH levels and hydrogen peroxide supplementation. New Microbiol. 33, 137–145 (2010).
Arciola, C. R. et al. Search for the insertion element IS256 within the ica locus of Staphylococcus epidermidis clinical isolates collected from biomaterial-associated infections. Biomaterials 25, 4117–4125 (2004).
Zhang, Y. Q. et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49, 1577–1593 (2003).
Acknowledgements
The contribution of Janni Kujala to the fluorescence microscopy experiments is greatly appreciated. We also acknowledge the financial support from Svenska Kulturfonden, Niilo V - Lauri N Santasalo (KAUTE) Foundation, Academy of Finland (BIOARMI project, decision 128870) and Drug Discovery and Chemical Biology (DDCB) network of Biocenter Finland.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Skogman, M., Vuorela, P. & Fallarero, A. Combining biofilm matrix measurements with biomass and viability assays in susceptibility assessments of antimicrobials against Staphylococcus aureus biofilms. J Antibiot 65, 453–459 (2012). https://doi.org/10.1038/ja.2012.49
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2012.49
Keywords
This article is cited by
-
The ribonuclease PNPase is a key regulator of biofilm formation in Listeria monocytogenes and affects invasion of host cells
npj Biofilms and Microbiomes (2023)
-
Janus nanoparticles targeting extracellular polymeric substance achieve flexible elimination of drug-resistant biofilms
Nature Communications (2023)
-
A systematic review of the use of bacteriophages for in vitro biofilm control
European Journal of Clinical Microbiology & Infectious Diseases (2023)
-
Antibacterial Effect of Phenolic Acids Derived from Rice Straw and in Combination with Antibiotics Against Escherichia coli
Applied Biochemistry and Biotechnology (2022)
-
Destruction of Staphylococcus aureus biofilms by combining an antibiotic with subtilisin A or calcium gluconate
Scientific Reports (2021)