Abstract
Strain BFLP-10T, isolated from faeces of wild long-snouted seahorses (Hippocampus guttulatus), is a Gram-negative, motile and facultatively anaerobic rod. This bacterium produces inhibitory activity against Vibrio species. Phylogenetic analysis based on 16S rRNA gene sequences showed that strain BFLP-10T was a member of the genus Vibrio and was most closely related to Vibrio owensii (99%), Vibrio communis (98.9%), Vibrio sagamiensis (98.9%) and Vibrio rotiferianus (98.4%). However, multilocus sequence analysis using gyrB, pyrH, recA and topA genes revealed low levels of sequence similarity (<91.2%) with these closely related species. In addition, strain BFLP-10T could be readily differentiated from other closely related species by several phenotypic properties and fatty acid profiles. The G+C content of the DNA was 45.6 mol%. On the basis of phenotypic, chemotaxonomic and phylogenetic data, strain BFLP-10T represents a novel species within the genus Vibrio, for which the name Vibrio inhibens sp. nov. is proposed. The type strain is BFLP-10T (=CECT 7692T=DSM 23440T).
Similar content being viewed by others
Introduction
The family Vibrionaceae currently comprises six validly published genera: Vibrio,1Photobacterium,2Salinivibrio,3Enterovibrio,4Grimontia5 and Aliivibrio.6Vibrio species are common inhabitants of aquatic environments, and they can be isolated under a wide range of salinity and temperature conditions from oysters, clams, mussels, and fish, as well as from sediment and plankton.7, 8
Some species have been reported to cause infections in humans and aquatic animals,9, 10 whereas a small number of other species have been used as probiotics in aquaculture.11, 12 In the present study, a Gram-negative bacterium (BFLP-10T), with inhibitory activity against Vibrio species, was isolated from faeces of wild long-snouted seahorses (H. guttulatus) captured in northwest Spain. The phylogenetic analysis based on the 16S rRNA gene of BFLP-10T indicated that it is closely related to members of the Harveyi clade. Thus, we characterize strain BFLP-10T and describe the identification of this novel species.
Materials and methods
Culture conditions
Strain BFLP-10T was isolated from faeces of wild long-snouted seahorses by using the standard dilution plating method on marine agar at 20 °C for 72 h. The strain was subcultured on the same medium at 22 °C for 24 h. Stock cultures were stored at −80 °C in marine broth with 30% (v/v) glycerol.
Physiological and biochemical characterization
Gram reaction was determined using the non-staining (KOH) method.13 Cell morphology and motility were studied using phase-contrast microscopy and electron microscopy as previously described by Herrera et al.14 NaCl growth tolerance and requirements were investigated by using nutrient broth (0.5% peptone from casein, 0.3% meat extract, 0.3% yeast extract, and adjusted to pH 7.2) supplemented with various concentrations of NaCl (0–15% at intervals of 1%). The pH range for growth was determined in marine broth that was adjusted to various pH values with acetic acid-sodium acetate (pH 4.0–4.5, 100 mM), MES (pH 5.0–6.0, 50 mM), MOPS (pH 6.5, 50 mM), Tris (pH 7.0–9.0, 50 mM) or CHES (pH 9.5–10.0, 50 mM). Growth temperature between 5 and 40 °C was tested in nutrient broth supplemented with 2% w/v NaCl for 48 h with shaking. Anaerobic growth was assessed at 22 °C in anaerobic chambers with an H2/CO2 atmosphere (BioMérieux, Marcy l'Etoile, France).
Catalase activity was determined by assessing bubble production in 3% (v/v) H2O2; oxidase activity was determined using 1% (w/v) tetramethyl-p-phenylenediamine as described by Lim et al.15 Some physiological characteristics were performed using API 20E, API 20NE and API ZYM (BioMérieux). Cells for inoculation of the strips were grown for 24 h at 22 °C on marine agar and the results were visually interpreted according to the manufacturer's instructions.
Antimicrobial activity was determined as described by Balcázar et al.16 Briefly, strain BFLP-10T was grown in 100 ml of marine broth without agitation at 22 °C for 48 h. After incubation, bacteria were removed by centrifugation (2000 g), and cell-free culture supernatant was recovered by passage through 0.22 μm pore size filters. The cell-free culture supernatant was adjusted to pH 6.5 with 5M NaOH to eliminate the inhibitory effects produced by organic acids. Moreover, sensitivity of cell-free culture supernatant to trypsin and proteinase K (Sigma Chemical Co., St Louis, MO, USA) at a final concentration of 1.0 mg ml−1 was also tested in buffers recommended by the supplier. Samples with and without enzymes were incubated at 37 °C for 2 h and residual activity was determined. To exclude potential inhibition by hydrogen peroxide, we added catalase (Sigma Chemical Co.) at a final concentration of 0.5 mg ml−1 and incubated at 37 °C for 30 min. All assays were independently repeated at least two times for reproducibility.
Five strains, Vibrio alginolyticus N26-1, Vibrio harveyi HT351, Vibrio ichthyoenteri HT21, Vibrio parahaemolyticus HT352 and Vibrio splendidus HT29, which have been isolated and identified in previous studies, were used as indicator bacteria.10, 16 Briefly, bacterial strains were grown in 5 ml of marine broth at 22 °C for 24 h. The cells were harvested by centrifugation (2000 g), washed twice with sterile saline solution and resuspended in 5 ml of the same solution. The bacterial suspensions were spread on marine agar plates. Four wells were made in each agar plate with a sterile Pasteur pipette, and cell-free culture supernatants (10 μl) from strain BFLP-10T were placed into each well. The plates were incubated aerobically at 22 °C for 24 h and then examined for zones of inhibition.
Genotypic characterization
Genomic DNA extraction, PCR amplification and sequencing of the 16S rRNA, DNA gyrase B subunit (gyrB), urydilate kinase (pyrH), recombination repair protein (recA) and topoisomerase I (topA), were carried out as described previously by Sawabe et al.17 and Balcázar et al.18 The sequences obtained were compared against the sequences available in the GenBank, EMBL and DDBJ databases obtained from the National Center for Biotechnology Information using the BLASTN.19 Phylogenetic analysis was performed using the software MEGA version 4.0 (Center for Evolutionary Medicine and Informatics, Tempe, AZ, USA) after multiple alignments of data by CLUSTAL X.20, 21 Distances (distance options according to the Kimura two-parameter model) and clustering with the neighbour-joining and maximum-parsimony methods were determined using bootstrap values based on 1000 replications.
For base composition analysis, DNA was prepared according to Chun & Goodfellow.22 The G+C content of the DNA was determined using the thermal denaturation method.23 DNA from Escherichia coli ATCC 11775T and Vibrio azureus LMG 25266T were used as a reference for determination of the thermal-melting profile (Tm).
Chemotaxonomic analysis
Whole-cell fatty acids from the isolate were extracted from biomass grown on tryptone soy agar (TSA) supplemented with 1.5% NaCl (w/v) and analysed according to the standard protocol of the Sherlock Microbial Identification System, version 4.5 (MIDI Inc., Newark, DE, USA).
Results and Discussion
Phenotypic characteristics
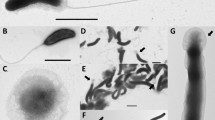
Phenotypically, strain BFLP-10T can be clearly assigned to the genus Vibrio, and belongs to the arginine-dihydrolase-negative, lysine-and ornithine-decarboxylase-positive species.24 Cells of strain BFLP-10T were slightly curved rods, Gram-negative, oxidase- and catalase-positive, motile and facultatively anaerobic. The novel strain also showed prolific growth on thiosulphate-citrate-bile salts-sucrose agar, forming green colonies. Strain BFLP-10T can be differentiated from related species on the basis of some biochemical properties such as the lack of mannose assimilation and sucrose fermentation. Other physiological characteristics of strain BFLP-10T are shown in Table 1 and also in the species description.
In addition, cell-free culture supernatant from strain BFLP-10T exhibited antibacterial activity against all indicator strains. Inhibitory activity of the cell-free culture supernatant was inactivated by enzyme treatment, which indicates that the inhibitory compound is proteinaceous. This action against closely related species provides evidence that the inhibitor involved could be a bacteriocin.25
Phylogenetic analysis
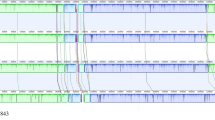
The 16S rRNA gene sequence of strain BFLP-10T was a continuous stretch of 1484 bp. In the neighbour-joining phylogenetic tree based on 16S rRNA gene sequences (Figure 1), strain BFLP-10T was closely related to V. owensii DY05T (99.0%), V. communis R-40496T (98.9%), V. sagamiensisT (98.9%) and V. rotiferianus LMG 21460T (98.4%). Similar results were obtained with the maximum-parsimony algorithm (Supplementary Information, Figure S1). Multilocus sequencing analysis of housekeeping genes has been used previously for inferring evolutionary relationships among members of the genus Vibrio.17, 26 In the present study, we used four housekeeping genes, gyrB, pyrH, recA and topA, in order to establish the taxonomic position of strain BFLP-10T. The phylogenetic trees based on gyrB, pyrH, recA and topA gene sequences revealed low levels of similarity (<91.2%) between strain BFLP-10T and the most closely related species (Figure 2). Previous studies have demonstrated that multilocus sequencing analysis has a higher discriminatory power than DNA–DNA hybridization.27, 28 In fact, Thompson et al.29 have reported that when two Vibrio strains share more than 95% similarity by multilocus sequencing analysis, they are the same species. These results demonstrate that strain BFLP-10T is distinct from any recognized species of the genus Vibrio.
Phylogenetic dendrogram of strain BFLP-10T with the most closely related Vibrio species, based on 16S rRNA gene sequences and constructed by the neighbour-joining method. Bootstrap percentages (>50%) based on 1000 replications are shown at branch nodes. Vibrio cholerae ATCC 14035T was used as an outgroup. Bar, 0.01 substitutions per nucleotide position.
Phylogenetic tree based on the concatenated sequences of the four housekeeping genes (2237 bp). This tree combines the results of both the neighbour-joining (NJ) and maximum-parsimony (MP) methods. The topology shown was obtained by using the NJ method. Bootstrap percentages based on 1000 replications are shown at branch nodes (NJ/MP). V. cholerae ATCC 14035T was used as an outgroup. Bar, 0.02 substitutions per nucleotide position.
Chemotaxonomic characteristics and DNA base composition
The major fatty acids in strain BFLP-10T were C15:0 iso 2-OH and/or C16:1ω7c (42.6%), C18:1ω7c (19.6%), C16:0 (16.2%) and C12:0 (3.8%), which comprise approximately 82.2% of the total fatty acid methyl esters detected. Fatty acids C15:0 iso 2-OH and/or C16:1ω7c, C16:0, C18:1ω7c, C14:0, C12:0 and C16:0 iso are typically the major fatty acids found in Vibrio species.30 However, strain BFLP-10T and most closely related-type strains, V. owensii, V. communis, V. sagamiensis and V. rotiferianus, could be clearly distinguished from each other based on the relative fatty acid concentration (Table 2). The DNA G+C content was calculated to be 45.6 mol%. This value is within the range for the genus Vibrio.31
Therefore, the phenotypic and genotypic properties of strain BFLP-10T support its description as a novel species within the genus Vibrio, for which the name V. inhibens sp. nov. is proposed.
Description of V. inhibens sp. nov.
V. inhibens (in.hi′bens. L. part. adj. inhibens, inhibiting). Cells are Gram-negative, motile, facultatively anaerobic and slightly curved rod-shaped (0.6 × 2.0–2.5 μm). Colonies on TSA supplemented with 1.5% (w/v) NaCl are cream coloured, circular and 1.5–2.0 mm in diameter. Growth occurs at NaCl concentrations between 1.0 and 8.0% (w/v), but not without NaCl or in the presence of >9.0% NaCl (w/v); grows at 10–35 °C (optimum 22 °C), but not at 5 and 40 °C; grows at pH 5.0–10.0 (optimum pH 7.0), but not at pH 4.5 or pH 10.5. Positive for catalase, oxidase, lysine-and ornithine-decarboxylase, indole production, nitrate reduction to nitrite, urease, aesculin, tryptophane deaminase, gelatine hydrolysis, N-acetyl-β-glucosamine, assimilation of D-glucose, D-mannitol and malate. Negative for acetoin production, arginine dihydrolase, production of H2S, assimilation of L-arabinose, D-mannose, D-maltose, potassium gluconate, caprate, citrate, adipate and phenyl-acetate. Acid is produced from amygdalin, D-glucose, D-fructose, D-mannitol, D-cellobiose, D-trehalose, amidon and glycogen but not from glycerol, erythritol, D-arabinose, L-arabinose, D-ribose, D-xylose, L-xylose, D-adonitol, methyl β-D-xylopyranoside, D-mannose, D-galactose, L-sorbose, L-rhamnose, dulcitol, inositol, D-sorbitol, methyl α-D-mannopyranoside, methyl α-D-glucopyranoside, arbutin, salicin, D-maltose, D-lactose, D-melibiose, D-saccharose, inulin, D-melezitose, D-raffinose, xylitol, gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol and L-arabitol. API ZYM tests show activities for alkaline phosphatase, β-galactosidase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase and naphthol-AS-BI-phosphohydrolase, but not for α-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase and α-fucosidase. The major fatty acids are C16:0, C12:0 and C18:1ω7c; smaller amounts of C14:0 and C12:0 3-OH are present. Undefined fatty acids are also observed, summed feature 2 (C14:0 3-OH and/or C16:1 iso I) and summed feature 3 (C15:0 iso 2-OH and/or C16:1ω7c ). The G+C content of the type strain is 45.6 mol% (Tm).
The type strain is BFLP-10T (=CECT 7692T=DSM 23440T), isolated from the faeces of wild seahorses captured in northwest Spain (Toralla, Galicia).
Accession numbers
The GenBank/EMBL/DDBJ accession numbers for the sequences of V. inhibens sp. nov. are FN687911 (16S rRNA gene), FR669655 (recA gene), HE588134 (gyrB gene), HE588135 (pyrH gene) and HE588136 (topA gene).
References
Baumann, P. & Schubert, R. H. W. in Bergey′s Manual of Systematic Bacteriology Vol. 1 (eds Krieg, N. R. & Holt, J. G.) 516–517 Williams & Wilkins: Baltimore, MD, (1984).
Baumann, P. & Baumann, L. in Bergey′s Manual of Systematic Bacteriology Vol. 1 (eds Krieg, N. R. & Holt, J. G.) 539–545 Williams & Wilkins: Baltimore, MD, (1984).
Mellado, E., Moore, E. R. B., Nieto, J. J. & Ventosa, A. Analysis of 16S rRNA gene sequences of Vibrio costicola strains: description of Salinivibrio costicola gen. nov., comb. nov. Int. J. Syst. Bacteriol. 46, 817–821 (1996).
Thompson, F. L. et al Enterovibrio norvegicus gen. nov., sp nov., isolated from the gut of turbot (Scophthalmus maximus) larvae: a new member of the family Vibrionaceae. Int. J. Syst. Evol. Microbiol. 52, 2015–2022 (2002).
Thompson, F. L., Hoste, B., Vandemeulebroecke, K. & Swings, J. Reclassification of Vibrio hollisae as Grimontia hollisae gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 53, 1615–1617 (2003).
Urbanczyk, H., Ast, J. C., Higgins, M. J., Carson, J. & Dunlap, P. V. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 57, 2823–2829 (2007).
Aznar, R., Ludwig, W., Amann, R. I. & Schleifer, K. H. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of Vibrio vulnificus with rRNA-targeted oligonucleotide probes. Int. J. Syst. Bacteriol 44, 330–337 (1994).
Thompson, F. L., Iida, T. & Swings, J. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431 (2004).
Morris, J. G. Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin. Infect. Dis 37, 272–280 (2003).
Balcázar, J. L., Gallo-Bueno, A., Planas, M. & Pintado, J. Isolation of Vibrio alginolyticus and Vibrio splendidus from cautive-bred seahorses with disease symptoms. Antonie van Leeuwenhoek 97, 207–210 (2010).
Balcázar, J. L., de Blas, I., Ruiz-Zarzuela, I., Cunningham, D., Vendrell, D. & Múzquiz, J. L. The role of probiotics in aquaculture. Vet. Microbiol. 114, 173–186 (2006).
Pérez, T. et al Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol 3, 355–360 (2010).
Buck, J. D. Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl. Environ. Microbiol 44, 992–993 (1982).
Herrera, J. J. R., Cabo, M. L., González, A., Pazos, I. & Pastoriza, L. Adhesion and detachment kinetics of several strains of Staphylococcus aureus subsp. aureus under three different experimental conditions. Food Microbiol. 24, 585–591 (2007).
Lim, J. M. et al Albimonas donghaensis gen. nov., sp. nov., a non-photosynthetic member of the class Alphaproteobacteria isolated from seawater. Int. J. Syst. Evol. Microbiol. 58, 282–285 (2008).
Balcázar, J. L., Loureiro, S., Da Silva, Y. J., Pintado, J. & Planas, M. Identification and characterization of bacteria with antibacterial activities isolated from seahorses (Hippocampus guttulatus). J. Antibiot. 63, 271–274 (2010).
Sawabe, T., Kita-Tsukamoto, K. & Thompson, F. L. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189, 7932–7936 (2007).
Balcázar, J. L., Pintado, J. & Planas, M. Bacillus galliciensis sp. nov., isolated from faeces of wild seahorses (Hippocampus guttulatus). Int. J. Syst. Evol. Microbiol. 60, 892–895 (2010).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignement aided by quality analysis tools. Nucleic Acid Res 25, 4876–4882 (1997).
Chun, J. & Goodfellow, M. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45, 240–245 (1995).
Mandel, M. & Marmur, J. Use of ultraviolet absorbance temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol 12B, 195–206 (1968).
Noguerola, I. & Blanch, A. R. Identification of Vibrio spp. with a set of dichotomous keys. J. Appl. Microbiol. 105, 175–185 (2008).
Shehane, S. D. & Sizemore, R. K. Isolation and preliminary characterization of bacteriocins produced by Vibrio vulnificus. J. Appl. Microbiol. 92, 322–328 (2002).
Thompson, F. L. et al Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71, 5107–5115 (2005).
Martens, M., Dawyndt, P., Coopman, R., Gillis, M., De Vos, P. & Willems, A. Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int. J. Syst. Evol. Microbiol. 58, 200–214 (2008).
Hoffmann, M., Monday, S. R., Fischer, M. & Brown, E. W. Genetic and phylogenetic evidence for misidentification of Vibrio species within the Harveyi clade. Lett. Appl. Microbiol. 54, 160–165 (2012).
Thompson, C. C. et al Genomic taxonomy of vibrios. BMC Evol. Biol 9, 258 (2009).
Thompson, F. L. & Swings, J. in The biology of vibrios eds F. L. Thompson, B. Austin, J. Swings 29–43 ASM Press: Washington, DC, (2006).
Farmer, J. J. in The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications eds A. Balows, H. G. Trüper, M. Dworkin, W. Harder, K. H. Schleifer2938–2951 Springer-Verlag: New York, NY, (1992).
Cano-Gómez, A., Goulden, E. F., Owens, L. & Høj, L. Vibrio owensii sp. nov., isolated from cultured crustaceans in Australia. FEMS Microbiol. Lett. 302, 175–181 (2010).
Chimetto, L. A. et al Vibrio communis sp. nov., isolated from the marine animals Mussismilia hispida, Phyllogorgia dilatata, Palythoa caribaeorum, Palythoa variabilis and Litopenaeus vannamei. Int. J. Syst. Evol. Microbiol. 61, 362–368 (2011).
Yoshizawa, S., Wada, M., Yokota, A. & Kogure, K. Vibrio sagamiensis sp. nov., luminous marine bacteria isolated from sea water. J. Gen. Appl. Microbiol 56, 499–507 (2010).
Acknowledgements
This study was funded by the Spanish Ministry of Science and Innovation (Projects CGL2005-05927-C03-01 and CGL2009-08386) and by the Regional Government Xunta de Galicia (09MDS022402PR). JLB was supported by a postdoctoral I3P contract from the Spanish National Research Council (CSIC), co-financed by the European Social Fund. We thank P Quintas, A Chamorro, M Cueto, S Otero and P Ruiz for skilful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Balcázar, J., Planas, M. & Pintado, J. Vibrio inhibens sp. nov., a novel bacterium with inhibitory activity against Vibrio species. J Antibiot 65, 301–305 (2012). https://doi.org/10.1038/ja.2012.22
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2012.22