Abstract
Objective:
Sensing of nutrients in the stomach is of crucial importance for the regulation of ingestive behavior especially in the context of metabolic dysfunctions such as obesity. Cells in the gastric mucosa with taste-signaling elements are considered as candidates for sensing the composition of ingested food and consequently modulate gastrointestinal processes. To assess whether obesity might have an impact on gastric chemosensory cells, gastric tissue samples from morbidly obese patients and normal-weight subjects were compared using a reverse transcriptase (RT)-PCR, qPCR and immunohistochemical approach.
Results:
Analysis of biopsy tissue samples from human stomach revealed that transcripts for the taste-signaling elements, including the receptor T1R3 involved in the reception of amino acids and carbohydrates, the fatty acid receptor GPR120, the G protein gustducin, the effector enzyme PLCβ2 and the ion channel TRPM5 are present in the human gastric mucosa and led to the visualization of candidate chemosensory cells in the stomach expressing gustatory marker molecules. RT-PCR and qPCR analyses indicated striking differences in the expression profiles of specimens from obese subjects compared with controls. For GPR120, gustducin, PLCβ2 and TRPM5 the expression levels were increased, whereas for T1R3 the level decreased. Using TRPM5 as an example, we found that the higher expression level was associated with a higher number of TRPM5 cells in gastric tissue samples from obese patients. This remarkable change was accompanied by an increased number of ghrelin-positive cells.
Conclusions:
Our findings argue for a relationship between the amount of food intake and/or the energy status and the number of candidate chemosensory cells in the gastric mucosa.
Similar content being viewed by others
Introduction
Endocrine and neuronal signals originating from the gastrointestinal (GI) tract directly control physiological processes, which affect energy homeostasis, food intake and satiety. The type and intensity of the signals strongly depend on the chemical composition of ingested food. Accordingly, an efficient chemosensory monitoring of the luminal content is an essential prerequisite for fine tuning not only the GI processes but also for regulating the feedback onto brain centers, which control food intake. An assessment of the luminal content is assumed to be accomplished by specialized chemosensory cells in the GI mucosa. Mounting evidence supports the view that GI mucosal cells with a molecular phenotype related to lingual taste receptor cells may operate as sensors for ingested food.1, 2, 3 Previous studies have mainly concentrated on the phenotypical characterization and topographical distribution of chemosensory cells in the small intestine, where the main resorption processes occur. Recent reports have provided first evidence for an interplay between a chemosensory registration of nutrients and the release of GI hormones.4, 5 In contrast, little is known about chemosensation in the stomach where digestive processes are initiated and major feedback signals originate. A few studies have shown that elements associated with taste transduction processes, such as the G protein gustducin, the key effector enzyme PLCβ2, the transient receptor potential ion channel TRPM5 and the G protein coupled receptor T1R3 are expressed in the gastric corpus mucosa of rodents and humans.6, 7, 8, 9, 10, 11 Moreover, it has been demonstrated that exogenous components elicit the release of ghrelin from gastric mucosal cells12 including bitter tastants,13 which suggests that a chemosensory registration of luminal constituents affects the secretion of hormones in the stomach. Because of the important implications of signals from the stomach for the regulation of central mechanisms governing food intake and energy homeostasis we investigated tissue samples from the human stomach for the appearance and distribution of chemosensory cells aiming for the important goal to unravel if the gastric chemosensory system may be affected under conditions of metabolic disorders as in morbid obesity.
Materials and methods
Human tissues
Human gastric tissue samples were taken from the gastric corpus of either normal-weight (body mass index 19–27 kg m−2) or morbidly obese subjects (body mass index 47–72 kg m−2) in the department for general, visceral and transplant surgery in the University Hospital in Tübingen and in the Clinic of surgery Munich-Bogenhausen. Specimens of morbidly obese patients derived from sleeve gastrectomies and endoscopic biopsies for routine diagnostic purposes. Among the normal-weight subjects, three participants were not suffering from cancer. Owing to the scarcity of endoscopy of non-obese healthy subjects, the majority of macroscopically normal gastric tissue of lean individuals was obtained from cancer patients who underwent either endoscopy for exclusion of cancerous alterations in the GI tract or gastric resections due to esophageal and pancreatic cancer. Exclusion criteria for all patients were type 2 diabetes, gastritis, instable body weight and infection with Helicobacter pylori. In total, 11 participating patients were morbidly obese (8 males and 3 females with a mean age of 44.5 years ranging from 26–58 years) and 11 subjects were non-obese (7 males and 4 females with a mean age of 52.1 years ranging from 25–74 years). After sampling of mucosal tissue, material was immediately transferred into a collection tube, frozen on dry ice and stored at −70 °C until use. For PCR analyses, gastric specimens of biopsies and surgical interventions were obtained within the greater curvature of the gastric corpus. As an internal control, from some patients gastric specimens were collected from adjacent positions to compare independent samples in parallel. For quantitative immunohistochemical analyses, larger biopsy samples (non-obese and obese), surgically resected specimens (non-obese) and gastric material from sleeve gastrectomies (obese) were used to ensure high histological quality. Sample collection and experiments were performed in accordance to the approval of the Ethics Commission Board of the University Hospital in Tübingen, Germany, and written informed consent was obtained from the participating patients.
RNA isolation, reverse transcriptase (RT)-PCR, qPCR, cloning and sequencing
Total RNA of frozen stomach specimens from normal-weight and obese subjects was prepared by the NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's protocol. To ensure the complete removal of DNA, a DNase digestion (DNase I, Life Technologies, Carlsbad, CA, USA) step was included. Subsequently, 0.8–1.2 μg total RNA was reversely transcribed using oligo(dT) primers and SuperScript III Reverse Transcriptase (RT) (Invitrogen, Carlsbad, CA, USA) followed by a conventional PCR amplification using High Fidelity PCR Enzyme Mix (Fermentas, St Leon-Rot, Germany). RNA integrity of each sample was controlled by the amplification of the housekeeping gene for the ribosomal protein L8 with intron spanning primers to verify the DNA removal. As a further control, RT-PCRs with RT samples were performed in which the RT had been omitted. For the amplification of gustatory-signaling elements, 1.2 μg of gastric cDNA from three normal-weight subjects was analyzed in independent PCR experiments. PCR amplification (25 μl) was carried out using 1 μl of the cDNA, 0.1 mM of each dNTP, 12.5 pmol of each primer and 0.75 U of the High Fidelity PCR Enzyme Mix in 1 × High Fidelity PCR Buffer with MgCl2. For amplification the following PCR cycling profiles were used. Profile 1: one cycle: 2 min at 94 °C; 20 cycles: 30 s at 94 °C, 40 s at 60 °C with −0.5 °C per cycle, 20 s at 72 °C; 20 cycles: 30 s at 94 °C, 30 s at 50 °C, 20 s at 72 °C and one cycle: 2 min at 72 °C. Profile 2: one cycle: 2 min at 94 °C; 34 cycles: 30 s at 94 °C, 1 min at 60 °C, 20 s at 72 °C and one cycle: 2 min at 72 °C. Exponential PCR amplification was performed to semi-quantify the expression levels of T1R3, GPR120, gustducin, PLCβ2 and TRPM5 using optimized number of amplification cycles to allow a comparison in a linear fashion. For semi-quantitative RT-PCR the above cycling profiles without the final elongation step at 72 °C for 2 min were used. A comparison of gastric specimens from obese and non-obese patients was performed in five pairs of semi-quantitative PCR series using 1.2 μg of reversely transcribed total RNA. To adjust the amount of cDNA, dilution series of the housekeeping gene L8 were amplified and equivalence of L8 bands was gauged on agarose gels. Equal intensity of L8 bands was assumed to reflect equal amounts of applied cDNA. Following adjustment of equal amounts of RNA, exponential amplification for T1R3, GPR120, PLCβ2 and TRPM5 was achieved during cycles 38–43, for gustducin during cycles 43–48.
For determination of quantitative changes in mRNA levels qPCR experiments were performed using the Light Cycler (Roche Diagnostics, Mannheim, Germany). The qPCR reaction mixture (20 μl) consisted of 2 × SYBR Green I Master Mix (Roche) or 2 × QuantiFast SYBR Green PCR Master Mix (Qiagen, Hilden, Germany) and primer sets. Relative amounts of transcripts for TRPM5, GPR120 and T1R3 were normalized to 18S rRNA quantification. The following qPCR protocol was used: 95 °C for 10 min, 95 °C for 15 s, 60 °C for 15 s, 72 °C for 20 s with 40 cycles, a melting step by slow heating from 65 to 95 °C with +0.5 °C per cycle and a final cooling down to 40 °C. Each assay included (in triplicate): for taste genes 50 ng of each tested cDNA, for 18S a 1:10 cDNA dilution, a non-template control reaction, a reaction of RT sample in which the RT had been omitted and a standard curve of four serial dilutions (in steps of fivefold) of a calibrator cDNA ranging from 250 to 2 ng for taste genes and ranging from 50 to 0.4 ng for 18S rRNA. LightCycler Software 3.5 (Roche Diagnostics) results were exported as tab-delimited text files and imported into Microsoft Excel for calculations of the expression ratios using the mean crossing points of target and reference genes from controls and samples. Efficiencies were acquired by the generation of standard curves of dilution series of a calibrator cDNA with each primer set and the given slopes in the LightCycler Software 3.5 (Roche Diagnostics).
For the amplification of gustatory-signaling elements the following primers were used: T1R3 primers, (nt 2169-2419 from GenBank accession number NM_152228; the expected size of PCR products, 251 bp) 5′-CAC ACG CTC CTG GGT CAG CTT CG-3′ and 5′-CCA GGA TGC CCA GGA CAC AGA G-3′; gustducin primers, 5′-CCA TTG TCC TGT TCC TCA ACA AA-3′ and 5′-GCC CAC AGT CTT TTA GAT TCT C-3′ (NM_001102386, nt 788-1057, 270 bp); PLCβ2 primers, 5′-CTT GGA GAG GCA CCA GGA GAA GC-3′ and 5′-CTT TGC TAT GAG TGG GTC CTG C-3′ (NM_004573, nt 3580-3832, 253 bp); TRPM5 primers, 5′-GGA GAC AGT CCA GAA GGA GAA CT-3′ and 5′-CTC GCC ACA GTG CTG AGA GCT C-3′ (NM_014555, nt 3171-3420, 250 bp; intron-spanning) and L8 primers, 5′-CAA GAA GAC CCG TGT GAA GC-3′ and 5′-GGC AGC AAT GAG ACC CAC TTT-3′ (NM_000973, nt 495-786, 292 bp, intron-spanning, 1490 bp genomic contamination). For 18S rRNA the quantitative qPCR primers were as follows: 5′-ATC AGA TAC CGT CGT AGT TC-3′ and 5′-CCA GAG TCT CGT TCG TTA T-3′ (NR_003286, nt 1055-1394, 340 bp), for TRPM5 the intron-spanning primers 5′-GAG CAA GAT GGA GAA GCG GAG GAG-3′ and 5′-GGA CAC GAG CAC CGA GCA GTA GTT-3′ (NM_014555, nt 3196-3360, 163 bp) and for GPR120 and T1R3 the used primers were listed above.
The amplified PCR products were cloned into pGEM-T vector (Promega, Medison, WI, USA) and validated by DNA sequencing using an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Partial sequences were matched to corresponding human full-length sequences by GenBank BLAST searches.
Immunohistochemistry
Frozen tissue was thawed in 1 × phosphate-buffered saline (PBS) (0.85% NaCl, 1.4 mM KH2PO4, 8 mM Na2HPO4 and pH 7.4) and fixed in 4% formalin/formaldehyde solution (Roti-Histofix, Roth, Karlsruhe, Germany) for 16 h at 4 °C followed by cryoprotection in 25% sucrose at 4 °C overnight. Subsequently, gastric tissue was embedded in Leica OCT cryocompound tissue freezing medium (Leica Microsystems, Bensheim, Germany) and quickly frozen on dry ice. Longitudinal sections (8 μm) were cut on a CM3000 cryostat (Leica Microsystems) and attached to Superfrost Plus microslides (Menzel Gläser, Braunschweig, Germany). For immunohistochemical staining sections were air-dried for 30 min and incubated with blocking solution (PBS with 10% normal goat serum (Dianova, Hamburg, Germany) or normal donkey serum (Dianova), 0.3% Triton X-100) for 30 min at room temperature. Then, the blocking solution was replaced by the primary antibody diluted in 1 × PBS containing 10% normal goat serum or normal donkey serum and 0.3% Triton X-100 at 4 °C overnight. Staining experiments were performed at the following dilutions: rabbit anti-TRPM59 1:600, rabbit anti-gustducin14 1: 200, rabbit anti-PLCβ2 1:300 (sc-206; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and goat anti-ghrelin (sc-10368; Santa Cruz Biotechnology) 1:300. For double staining the dilutions were rabbit anti-PLCβ2 1:300, rabbit anti-TRPM5 1:400 and goat anti-ghrelin 1:500, respectively, and normal donkey serum was used instead of normal goat serum. In control experiments, the primary antibody was omitted on consecutive tissue sections. After washing with PBS, the tissue sections were incubated with goat anti-rabbit Alexa 488-conjugated and/or donkey anti-goat Alexa 568-conjugated secondary antibody (Invitrogen, Karlsruhe, Germany; 1:500 in blocking solution) for 2 h at room temperature. Then, slides were washed with PBS. To visualize nuclei, 4′,6-diamidino-2-phenylindole (DAPI)-containing solution (1 μg ml−1 in PBS) was added to the tissue sections. For double staining sections were counterstained with TOTO-3 (Invitrogen). After rinsing the tissue sections in PBS, the sections were embedded in PBS/glycerol (1:3) or Mowiol (Roth) and coverslips sealed with nail polish.
Microscopy and imaging
Immunofluorescence was examined and documented with a Zeiss Axiophot microscope (Carl Zeiss MicroImaging, Göttingen, Germany) equipped with a SensiCam CCD camera (PCO Computer Optics, Kelheim, Germany) and double labeling by confocal microscopy using a Zeiss LSM 510 META system (Zeiss, Jena, Germany). Images were arranged and adjusted for contrast in PowerPoint (Microsoft) and Adobe Photoshop (Adobe Systems, San Jose, CA, USA).
Cell quantification
Cell counting was conducted adapting a procedure from Tanaka-Shintani and Watanabe15. In brief, random sampling fields of human gastric corpus mucosae of equal thickness were selected above the lamina muscularis where putative chemosensory cells and ghrelin-secreting cells mainly reside. To determine the number of TRPM5- and ghrelin-positive cells, microscopic digital images of four to five consecutive sections were acquired by defining sampling fields of 370 × 500 μm2 in basal zones of the gastric mucosa, which contained immunoreactive cells. Cell counting was performed in a blind manner, and the average cell counts of immunopositive cells per field were determined. Immunoreactive cells were only counted when the nuclei were clearly visible by DAPI staining. To define the percentage of immunolabeled cells per field, the total number of DAPI-labeled nuclei was counted.
Statistical analysis
For the quantification of relative changes in mRNA expression levels, data (in triplicate) were expressed as mean fold differences±s.e.m. compared with that of controls, with values=1 representing the baseline level corresponding to no relative difference in expression levels. The formula used to calculate the n-fold difference of mRNA expression levels of target genes relative to that of reference genes was as follows: ratio=(Etarget)ΔCt target(control–sample)÷(Eref)ΔCt ref (control–sample). For cell quantification, values are given as mean±s.e.m. for the quantification of four to five consecutive sections. Significant differences between the groups were analyzed by the unpaired t-test with GraphPad Prism (Graphpad Software, www.graphpad.com). Statistical significance was set at P<0.05.
Results
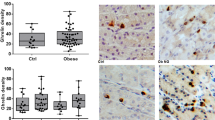
For the initiation and regulation of complex digestive processes, the stomach strongly relies on the capacity to sense the composition of the ingested food. In the mucosa of the GI tract, cells which express gustatory elements are considered as candidate chemosensors. By means of RT-PCR experiments we found transcripts for gustducin, PLCβ2 and TRPM5 in biopsies from human gastric corpus tissue (data not shown) confirming a previous report about the expression of gustatory-signaling elements in the human stomach.10 To visualize the candidate chemosensory cells within the gastric mucosa, which express taste-signaling elements, immunohistochemical analyses were performed. Tissue sections through the human gastric corpus mucosa were assessed using specific antibodies. Analyses with a TRPM5 antibody led to the labeling of either round or elongated gastric mucosal cells, which were located in the lower half of the human oxyntic glands (Figure 1a). No immunoreactivity was observed in the absence of the primary antibody (Figure 1b). TRPM5-immunoreactive cells extended their processes to either the apical or the basolateral site of oxyntic glands (Figures 1c and d). Using an antibody specific for PLCβ2 resulted in a weaker and more punctuated staining of similar cell types; a gustducin antibody labeled only few cells (data not shown).
Detection and localization of TRPM5 protein in the human gastric mucosa. (a) Immunohistochemistry using an antibody specific for TRPM5 revealed immunoreactive cells homogeneously distributed along glandular annuli in the basal half of the human corpus mucosa. (b) In the absence of the primary antibody no staining was evident. Dotted lines in (a) and (b) encircle gastric rosettes of consecutive sections. (c, d) TRPM5-immunoreactive cells displayed long cytoplasmic processes oriented to the apical (c) and/or basolateral (d) site of oxyntic glands. Cell nuclei were stained with DAPI. Scale bar (a–d) 10 μm.
To approach the question whether the energy status, especially obesity, may affect the number and distribution of putative chemosensory cells in the stomach mucosa, samples of gastric tissue obtained from morbidly obese patients were investigated; for comparison, samples from non-obese individuals were processed in parallel. First, semi-quantitative RT-PCR experiments for the taste-signaling elements gustducin, PLCβ2 and TRPM5 were performed. A comparison with samples from normal-weight individuals revealed that the bands for the analyzed genes were much stronger for gastric tissue from obese patients (Figure 2a; n=5, each). In addition, receptor genes were assessed, which may be relevant for sensing nutrients. T1R3 was selected as a possible chemosensor for both sugars and amino acids16, 17, 18 and GPR120 as a receptor for medium and long-chain fatty acids.19, 20, 21, 22 PCR experiments with cDNA from gastric corpus tissue from obese and normal-weight patients resulted in amplification products for both T1R3 and GPR120. For T1R3 the band was apparently weaker for gastric tissue from obese patients, conversely, for GPR120 the band was stronger (Figure 2b; n=5, each). To quantify the relative amounts of gene expression, mRNA levels for TRPM5, GPR120 and T1R3 were measured by real time PCR in morbidly obese and non-obese patients and expressed as fold differences of expression levels in normal-weight subjects relative to obese patients. In morbidly obese patients the amount of TRPM5 and GPR120 mRNA was 3.14-fold (P=0.0001) and 3.85-fold (P=0.0028) higher than in normal weight subjects, whereas the amount of T1R3 mRNA was 0.41-fold (P=0.0086) lower compared with controls (Figure 2c; n=4, each). These results indicate an elevated level of mRNA for taste-signaling elements and GPR120 and a lower level of mRNA for T1R3 in the gastric mucosa of obese patients.
Semi-quantitative RT-PCR and qPCR analyses of taste-signaling elements and candidate nutrient receptors comparing tissue samples from obese and normal-weight individuals. (a) For α-gustducin (270 bp), PLCβ2 (253 bp) and TRPM5 (250 bp) stronger bands were generated using cDNA from obese gastric tissue compared with band intensities obtained with cDNA from non-obese gastric tissue. (b) For T1R3—the candidate receptor for carbohydates and/or amino acids—stronger bands were obtained in gastric tissue samples from normal-weight individuals, whereas only traces of T1R3 RNA were detectable in obese tissue samples. In contrast, analyses with cDNA from normal-weight gastric tissue yielded only minor amounts of amplification products for GPR120—a candidate fatty acid receptor—compared with experiments using cDNA from obese gastric tissue. The analysis of band intensities for L8 (292 bp) served as a control; the comparison revealed similar results for the housekeeping gene indicative of equal template concentrations of the used templates. (c) Expression of TRPM5 and GPR120 mRNA of morbidly obese patients was markedly higher than that of controls as revealed by qPCR. In obese subjects, T1R3 was significantly expressed at lower levels. Data are expressed as mean fold differences±s.e.m., with values=1 representing the baseline level corresponding to no relative changes. Dashed line denotes baseline levels. Data were generated in triplicate, with n=4 subjects in each group. Statistically significant results determined by the unpaired t-test are indicated by *P<0.05; **P<0.005 against baseline levels. Note that error bars reflect strong interindividual differences.
The higher amount of mRNA for gustducin, PLCβ2, TRPM5 and GPR120 in obese gastric tissue could either be due to higher mRNA levels in individual cells or due to an increased number of cells expressing the signaling elements. To investigate the cell numbers, candidate chemosensory cells in the gastric mucosa were visualized by means of immunohistochemical approaches using the TRPM5 antibody. A typical result of these experiments is depicted in Figures 3a and b. It is immediately obvious that the number of TRPM5-immunopositive cells is higher in tissue samples from obese patients (Figure 3a) compared with tissue samples from normal-weight individuals (Figure 3b). To substantiate this observation, TRPM5-immunoreactive cells and DAPI-stained cell nuclei were counted on serial sections by defining a mucosal field of 370 × 500 μm2. Squares of counting fields were positioned in the upper half of the gastric mucosa above the lamina muscularis where TRPM5-positive cells are mainly distributed. The results are documented in Figure 3c. The average number of TRPM5-positive cells per field in the corpus of obese and normal-weight subjects was determined as 37.84±12.65 and 20.75±13.13 (n=7, obese; n=6, non-obese; P=0.0232), respectively (Figure 3c). To define the percentage of immunolabeled cells per field, the total number of DAPI-labeled nuclei was counted. By DAPI staining the average number of the total number of cells in obese and normal-weight patients was calculated as 722.49±148.02 and 718.49±95.80, respectively. Thus, the number of TRPM5-immunoreactive cells accounted for 5.35% of gastric mucosal cells in the basal half in obese subjects, and 2.99% in non-obese subjects. These findings indicate that the higher mRNA level for TRPM5 in tissue samples of obese patients associate with a higher number of TRPM5-positive cells in the gastric mucosa.
Distribution pattern and quantitative evaluation of TRPM5-immunoreactive cells in human gastric mucosa samples of obese and non-obese individuals. (a, b) Longitudinal sections of the human gastric mucosa immunostained for TRPM5 as a representative marker for the visualization of candidate chemosensory cells in the stomach. TRPM5-positive cells were apparently more frequently found on sections through the gastric mucosa of obese patients (a) than on gastric sections from normal-weight individuals (b). Cell nuclei were stained with DAPI. Scale bar (a, b), 50 μm. (c) Immunoreactive cells for TRPM5 were counted in a unit area of 370 × 500 μm2 on four to five consecutive longitudinal sections through the gastric mucosa. Mean numbers (±s.e.m.) of immunopositive cells in obese (hatched) and non-obese (dotted) subjects. Cell densities are expressed as labeled cell numbers per unit area. *P<0.05 compared with lean subjects.
It has recently been shown that in the murine stomach a population of candidate chemosensory cells also express the orexigenic hormone ghrelin.11 To assess whether in the human stomach candidate chemosensory cells may represent ghrelin-producing cells, double-labeling experiments were performed using TRPM5 and PLCβ2 antibodies. It was found that in the human oxyntic mucosa the vast majority of ghrelin-immunoreactive cells did not express TRPM5, but instead ghrelin-positive cells were frequently found in very close association with cells expressing TRPM5 (Figure 4a); only very few double-labeled cells were observed (data not shown). Double-labeling experiments with antibodies against PLCβ2 and ghrelin also revealed no overlap for the vast majority of cells (Figure 4b). Because of a close contact between putative chemosensory and ghrelin-secreting cells, we reasoned that the population of ghrelin-producing cells may also be increased in tissue from obese patients. Determining the number of ghrelin cells on tissue sections through the oxyntic mucosa of obese and non-obese subjects revealed a higher density of ghrelin cells in samples from obese subjects (Figure 5a) compared with controls (Figure 5b). The average number of ghrelin cells per field in samples from obese and non-obese subjects was 18.29±6.80 and 10.41±2.74 (n=7, obese; n=5, non-obese; P=0.0164), respectively (Figure 5c). These findings imply that gastric tissue of morbidly obese subjects is endowed with a higher number of both putative chemosensory cells and endocrine cells producing the orexigenic hormone ghrelin.
Localization and quantitative evaluation of ghrelin-immunoreactive cells in the human gastric mucosa. (a, b) Double-labeling experiments using antibodies against ghrelin (red) and TRPM5 (a, green) and PLCβ2 (b, green), respectively, revealed that ghrelin-releasing cells do not overlap with cells expressing the analyzed taste signaling molecules. In some cases ghrelin-positive cells were found to be in immediate proximity to cells immunoreactive for chemosensory elements. (a) Confocal image exemplarily depicting a ghrelin-positive cell (red) closely associated with a TRPM5-positive cell (green). Sections were counterstained with TOTO-3. Scale bar (a, b), 5 μm.
Distribution pattern and localization of ghrelin-immunoreactive cells in the human gastric mucosa. Ghrelin-positive endocrine cells appeared more frequent on sections through the gastric mucosa of obese patients (a) relative to ghrelin-immunoreactive cells on gastric sections from normal-weight individuals (b). Sections were counterstained with DAPI. Scale bar (a, b), 50 μm. (c) Immunoreactive cells for ghrelin were counted in a unit area of 370 × 500 μm2 on four to five consecutive longitudinal sections through the gastric mucosa. Mean numbers (±s.e.m.) of immunopositive cells in obese (hatched) and non-obese (dotted) subjects. Cell densities are expressed as labeled cell numbers per unit area. *P<0.05 compared with lean subjects.
Discussion
The results of this study indicate that the human gastric mucosa includes distinct cell types, which share some molecular features with lingual taste sensory cells. Based on the view that TRPM5 is accepted as a suitable marker for solitary chemosensory cells9, 23 a substantial number of candidate chemosensory cells was visualized in tissue samples from human gastric mucosa; the number in the lower half of the human corpus oxyntic glands accounted for ∼3% of the mucosal cells. Many of the TRPM5-positive cells were found to extend long cytoplasmic processes, which occurred to be directed toward the gastric lumen. This morphological feature suggests that the putative sensory cells may have direct contact to the luminal content and may be capable of sensing constituents of the ingested food. It is currently unclear which nutrient receptors are present in these candidate chemosensory cells, but RT-PCR experiments have shown that T1R3 as well as the fatty acid receptor GPR120 are expressed in human gastric mucosa. Evidence for the expression of T1R3 in the human upper GI tract was recently also found by Young et al.10 It is unclear how these candidate sensory cells are integrated in the complex physiology of the GI tract, that is, how they convey sensory information onto effector systems; however, it is conceivable that they may transmit their information onto neighboring nerve fibers of the enteric nervous system or onto adjacent endocrine cells via paracrine mechanisms. Alternatively, one could imagine that they do have endocrine function themselves and thus represent enteroendocrine cells with sensory capacity.
One of the major finding of this study was the substantial change in the expression level for a variety of chemosensory elements in the gastric mucosa of obese patients. These changes could be part of adaptive response mechanisms adjusting the GI system to a sustained positive energy balance. Whereas for most elements a higher expression level was observed, for the receptor T1R3, which is a constituent of sugar and amino acid receptors, the level was decreased. These findings are in line with a recent report demonstrating that in the lingual gustatory system of obese rats the expression level of T1R3 was reduced.24 It is conceivable that such a downregulation of T1R3 expression in the gastric and lingual system might be due to desensitization of the cells as a consequence of a sustained access to carbohydrate and/or protein breakdown products, but the finding that for the fatty acid receptor GRP120 as well as for the gustatory-signaling elements the expression levels were increased indicate that in obese gastric mucosa chemosensory elements are subject to different regulatory mechanisms. The reciprocal change of the mRNA levels for T1R3 and GPR120 might be an indication that the receptors are expressed in distinct cell populations.
Using TRPM5 as a marker for solitary chemosensory cells9, 23 our analyses revealed that the elevated expression level coincides with a significantly higher number and density of candidate chemosensory cells in the gastric mucosa from obese patients. It is unclear whether this phenomenon is one of the causal parameters for the disease or a consequence of the disorder, however, the changes in obese patients suggest that chemosensory cells are part of the complex regulation mechanisms involved in controling energy homeostasis. Whereas in the murine stomach some of the candidate chemosensory cells apparently also express the orexigenic hormone ghrelin, suggesting an immediate linkage between registration of nutritive signals and ghrelin release,15 in the human gastric mucosa the majority of cells expressing gustatory-signaling elements did not express ghrelin. This observation may suggest distinct features of ghrelin cells in different species. The adjacent localization of candidate chemosensory cells and ghrelin cells might indicate a paracrine cell–cell communication. In view of the finding that, in contrast to mice, in the human mucosa ghrelin and chemosensory elements are expressed in distinct cell types, the decrease in T1R3 mRNA concentration is not contradictory to the concomitant increase of ghrelin cells. An increased number and density of ghrelin cells in gastric biopsies from obese patients is in line with a previous report, indicating a trend toward an increased density of gastric ghrelin-positive cells in patients with a high body mass index.25 A higher density of ghrelin cells in obese gastric mucosa is remarkable because a decreased level of circulating ghrelin was found in obese patients,26 suggesting hypoactivity of ghrelin-secreting cells under these conditions.25 Whether such a hypoactivity of ghrelin cells could be due to inhibitory signals conveyed from chemosensory cells in response to the presence of nutrients needs further investigation.
Accession codes
References
Rozengurt N, Wu SV, Chen MC, Huang C, Sternini C, Rozengurt E . Colocalization of the α-subunit of gustducin with PYY and GLP-1 in L cells of human colon. Am J Physiol Gastrointest Liver Physiol 2006; 291: G792–G802.
Kokrashvili Z, Mosinger B, Margolskee RF . Taste signaling elements expressed in gut enteroendocrine cells regulate nutrient-responsive secretion of gut hormones. Am J Clin Nutr 2009; 90: 822S–825S.
Sbarbati A, Bramanti P, Benati D, Merigo F . The diffuse chemosensory system: exploring the iceberg toward the definition of functional roles. Prog Neurobiol 2010; 91: 77–89.
Jang HJ, Kokrashvili Z, Theodorakis MJ, Carlson OD, Kim BJ, Zhou J et al. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc Natl Acad Sci USA 2007; 104: 15069–15074.
Margolskee RF, Dyer J, Kokrashvili Z, Salmon KS, Ilegems E, Daly K et al. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc Natl Acad SCI USA 2007; 104: 15075–15080.
Höfer D, Püschel B, Drenckhahn D . Taste receptor-like cells in the rat gut identified by expression of á-gustducin. Proc Natl Acad Sci USA 1996; 93: 6631–6634.
Bezençon C, le Coutre J, Damak S . Taste-signaling proteins are coexpressed in solitary intestinal epithelial cells. Chem Senses 2007; 32: 41–49.
Hass N, Schwarzenbacher K, Breer H . A cluster of gustducin-expressing cells in the mouse stomach associated with two distinct populations of enteroendocrine cells. Histochem Cell Biol 2007; 128: 457–471.
Kaske S, Krasteva G, Konig P, Kummer W, Hofmann T, Gudermann T et al. TRPM5, a taste-signaling transient receptor potential ion-channel, is a ubiquitous signaling component in chemosensory cells. BMC Neurosci 2007; 8: 49.
Young RL, Sutherland K, Pezos N, Brierley SM, Horowitz M, Rayner CK et al. Expression of taste molecules in the upper gastrointestinal tract in humans with and without type 2 diabetes. Gut 2008; 58: 337–346.
Hass N, Schwarzenbacher K, Breer H . T1R3 is expressed in brush cells and ghrelin-producing cells of murine stomach. Cell Tissue Res 2010; 339: 493–504.
Yang GK, Yip L, Fredholm BB, Kieffer TJ, Kwok YN . Involvement of adenosine signaling in controlling the release of ghrelin from the mouse stomach. J Pharmacol Exp Ther 2011; 336: 77–86.
Janssen S, Laermans J, Verhulst P-J, Thijs T, Tack J, Depoortere I . Bitter taste receptors and á-gustducin regulate the secretion of ghrelin with functional effects on food intake and gastric emptying. Proc Natl Acad Sci USA 2011; 108: 2094–2099.
Widmayer P, Breer H, Hass N . Candidate chemosensory cells in the porcine stomach. Histochem Cell Biol 2011; 136: 37–45.
Tanaka-Shintani M, Watanabe M . Distribution of ghrelin-immunoreactive cells in human gastric mucosa: comparison with that of parietal cells. J Gastroenterol 2005; 40: 345–349.
Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS . Mammalian sweet taste receptors. Cell 2001; 106: 381–390.
Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ et al. An amino-acid taste receptor. Nature 2002; 416: 199–202.
Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E . Human receptors for sweet and umami taste. Proc Natl Acad Sci USA 2002; 99: 4692–4696.
Cartoni C, Yasumatsu K, Ohkuri T, Shigemura N, Yoshida R, Godinot N et al. Taste preference for fatty acids is mediated by GPR40 and GPR120. J Neurosci 2010; 30: 8376–8382.
Matsumura S, Eguchi A, Mizushige T, Kitabayashi N, Tsuzuki S, Inoue K et al. Colocalization of GPR120 with phospholipase-Câ2 and á-gustducin in the taste bud cells in mice. Neurosci Lett 2009; 450: 186–190.
Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010; 142: 687–698.
Montmayeur JP, Fenech C, Kusumakshi S, Laugerette F, Liu Z, Wiencis A et al. Screening for G-protein-coupled receptors expressed in mouse taste papillae. Flavour Fragr J 2011; 26: 223–230.
Hansen A, Finger TE . Is TrpM5 a reliable marker for chemosensory cells? Multiple types of microvillous cells in the main olfactory epithelium of mice. BMC Neurosci 2008; 9: 115.
Chen K, Yan J, Suo Y, Li J, Wang Q, Lv B . Nutritional status alters saccharin intake and sweet receptor mRNA expression in rat taste buds. Brain Res 2010; 1325: 53–62.
Liew PL, Lee WJ, Lee YC, Chen WY . Gastric ghrelin expression associated with Helicobacter pylori infection and chronic gastritis in obese patients. Obes Surg 2006; 16: 612–619.
Tschöp M, Weyer C, Tataranni PA, Devanarayan V, Ravussin E, Heiman ML . Circulating ghrelin levels are decreased in human obesity. Diabetes 2001; 50: 707–709.
Acknowledgements
We are grateful to Nicole Hass and Karin Schwarzenbacher for their valuable contributions to this study and thank Kerstin Bach for excellent technical assistance. We express our gratitude to T Gudermann and V Chubanov for generously providing the TRPM5 antibody. This work was supported by the ‘Kompetenznetz Adipositas (Competence Network of Obesity)’, research focus ‘Obesity and the GI tract’, funded by the Federal Ministry of Education and Research, Germany (no. 01GI0843).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Widmayer, P., Küper, M., Kramer, M. et al. Altered expression of gustatory-signaling elements in gastric tissue of morbidly obese patients. Int J Obes 36, 1353–1359 (2012). https://doi.org/10.1038/ijo.2011.216
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ijo.2011.216
Keywords
This article is cited by
-
Sweet taste receptors play roles in artificial sweetener-induced enhanced urine output in mice
npj Science of Food (2024)
-
Altered expression levels of TAS1R2 and TAS1R3 genes among SARS-CoV-2 variants of concerns
Molecular Biology Reports (2023)
-
Fat Taste Nerves and Their Function in Food Intake Regulation
Current Oral Health Reports (2022)
-
Association of TRPM5 Asn235Ser Polymorphism and Trace Elements/Minerals in Chronic Gastritis Patients: a Case-Control Study
Biological Trace Element Research (2022)
-
Effect of Endoscopic Gastroplication on the Genome-Wide Transcriptome in the Upper Gastrointestinal Tract
Obesity Surgery (2017)