Abstract
The preservation of biodiversity requires an understanding of the maintenance of its components, including genetic diversity. Effective population size determines the amount of genetic variance maintained in populations, but its estimation can be complex, especially when populations are interconnected in a metapopulation. Theory predicts that the effective size of a metapopulation (meta-Ne) can be decreased or increased by population subdivision, but little empirical work has evaluated these predictions. Here, we use neutral genetic markers and simulations to estimate the effective size of a putative metapopulation in Atlantic salmon (Salmo salar). For a weakly structured set of rivers, we find that meta-Ne is similar to the sum of local deme sizes, whereas higher genetic differentiation among demes dramatically reduces meta-Ne estimates. Interdemic demographic processes, such as asymmetrical gene flow, may explain this pattern. However, simulations also suggest that unrecognized population subdivision can also introduce downward bias into empirical estimation, emphasizing the importance of identifying the proper scale of distinct demographic and genetic processes. Under natural patterns of connectivity, evolutionary potential may generally be maintained at higher levels than the local population, with implications for conservation given ongoing species declines and habitat fragmentation.
Similar content being viewed by others
Introduction
Preserving biodiversity requires an understanding of the distribution and maintenance of its components, including genetic variation. The concept of effective population size (Ne) was developed by Wright (1931) to describe the behaviour of the neutral genetic variation of populations in relation to a theoretically ideal population. On the time scales of relevance to contemporary evolution and conservation, Ne is principally shaped by demography, mating system and life history (Caballero, 1994; Frankham, 1995; Gaggiotti and Vetter, 1999). A recent increase in interest in the estimation of contemporary Ne (for example, Schwartz et al., 2007; Palstra and Ruzzante, 2008; Waples and Do, 2010) has come with the realization that its conceptual simplicity is often mirrored by the high complexity of its empirical estimation (Wright, 1938; Nunney and Elam, 1994; Engen et al., 2005). Spatial segregation, population connectivity and temporal stratification (that is, age structure) can all complicate the behaviour and estimation of effective population size (for example, Jorde and Ryman, 1995; Waples, 2010; Hare et al., 2011). The effects of incoming gene flow on local Ne can be multifarious, depending on its relative magnitude, directional symmetry and the time frame of interest (Wang and Whitlock, 2003; Fraser et al., 2007; Palstra and Ruzzante, 2008).
Ne can also be considered from the relatively higher hierarchical level of a ‘metapopulation’ (Levins, 1969) or ‘subdivided population’ (Wright, 1969). At this level, the focus is on the effective size of an assemblage of populations interconnected by gene flow (hereafter referred to as meta-Ne; Fraser et al., 2007). This parameter has seen considerable theoretical treatment, starting with Wright (1943), who showed that under the restricted assumptions of his island model (that is, all demes of equal size and experiencing exactly the same level of symmetrical gene flow), population subdivision increases meta-Ne beyond the sum of local or deme effective sizes (ΣNe(s)). Subsequent authors have relaxed many of the assumptions inherent to this model (for example, Li, 1955; Caballero and Hill, 1992; Wang, 1996; Whitlock and Barton, 1997; Nunney, 1999; Tufto and Hindar, 2003; see Wang and Caballero (1999) and Whitlock (2004) for reviews) and have concluded that meta-Ne will only be larger than ΣNe(s) when density regulation acts locally and constrains reproductive variance (cf. Nunney, 1999; Whitlock, 2004). Conversely, factors enhancing overall variance in vital rates, such as unequal or asymmetrical deme contributions or extinction–recolonization dynamics, will reduce the effective size of a metapopulation. These reductions in meta-Ne can also be generated by directional abiological factors, such as oceanic currents, by introducing additional skew in the propagule contributions of individual demes (Wares and Pringle, 2008).
Population subdivision may thus generally lead to reductions in meta-Ne relative to ΣNe(s), yet this topic has seen little empirical study. For instance, Sugg et al. (1996) reported that population subdivision increased Ne in prairie dogs (but see Nunney (1999)). Also, asymmetrical gene flow among demes does not necessarily imply reduced meta-Ne. Morrissey and de Kerckhove (2009) showed that under conditions of hierarchical spatial structuring in dendritic systems, such as often seen in freshwater, asymmetrical gene flow can maintain high genetic diversity and thereby lead to high meta-Ne. Studies on anadromous salmonid fish (for example, Hindar et al., 2004; Fraser et al., 2007; Kuparinen et al., 2010), however, report that metapopulation effective size can be much reduced compared with the sum of local deme effective sizes. Using the approach of Tufto and Hindar (2003), Kuparinen et al. (2010) found that meta-Ne was mainly determined by the population with the highest emigration rate (see also Hindar et al. (2004)). In conclusion, the limited empirical work published to date provides contrasting results on the consequences of population subdivision for effective population size, and more empirical study is needed to better understand the conditions under which meta-Ne is expected to be larger or smaller than ΣNe(s).
Here, we focus on the effective size of a putative metapopulation in a widespread anadromous salmonid fish, Atlantic salmon (Salmo salar). The life history of Atlantic salmon makes this species suitable for metapopulation genetic studies. First, Atlantic salmon are generally spatially segregated into different freshwater systems, yet remain connected through the occasional exchange of migrant individuals that ‘stray’ into non-natal watersheds (Stabell, 1984; Hendry et al., 2004). Second, the species has been subject to extensive genetic study (Verspoor et al., 2008) and its spatial and temporal genetic structures are relatively well documented (for example, King et al., 2001; Dionne et al., 2008; Palstra and Ruzzante, 2010). Finally, small populations of Atlantic salmon often maintain surprisingly high levels of genetic diversity, suggesting that gene flow is not negligible and might often be asymmetrical (for example, Østergaard et al., 2003; Consuegra et al., 2005; Hansen et al., 2007; Palstra et al., 2007).
Our objective is twofold: we first estimate meta-Ne empirically in a putative metapopulation of Atlantic salmon in Newfoundland, Canada. We do this in two ways: first, by pooling river-specific samples into a single sample pool and considering the entire system as a single (though admittedly subdivided) population; second, we employ a model that requires knowledge of Ne of individual subpopulations (Ne(s)) and of migration rates among them. This latter model was also used to address our second objective: to determine the combinations of demographic factors (for example, unequal population sizes or strong gene flow asymmetries) that facilitate congruence between the two estimation methods, and are thus likely to be important in shaping meta-Ne. We, thus, explicitly assume that the meta-Ne estimate obtained with the first method is a reliable reference point for making demographic inferences in the second method. Additional simulations support our finding that population subdivision can reduce the effective size of a metapopulation, but that this reduction is most plausible under negatively density-dependent dispersal. Importantly, explorations of bias suggest that population subdivision itself, when ignored, may downwardly bias the empirical estimation of meta-Ne and we urge further theoretical and methodological developments.
Materials and methods
Study system
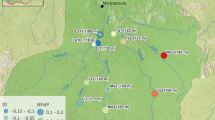
This study focuses on a set of 12 rivers inhabited by Atlantic salmon on the southwest coast of Newfoundland, Canada (Figure 1). These rivers flow into the Bay of St George and are hereafter called ‘the study system’. The river mouths are all located in relatively close proximity (within a 50-km stretch of coastline) but are relatively isolated from rivers outside the study system (by at least 60 km). These rivers lack the extensive lacustrine habitat typical of Newfoundland watersheds (see O’Connell and Ash (1993)) and contain salmon with a regionally distinct life history, comprising a relatively high proportion of multisea-winter anadromous salmon (that is, individuals that spend more than a single year at sea before returning to spawn in freshwater; O’Connell et al., 2006). Population census information is unavailable for five rivers, three of which are very small and may not sustain spawning salmon populations (B Dempson, personal communication). Further details on life history and regional habitat are given in Table 1 and Figure 1.
Molecular genetic analyses
Archived scale samples (N=395) were collected non-lethally from anadromous adult salmon in five waterways in the study system (all river mouths located within 50 km from each other), between 1973 and 1994. Contemporary samples from these rivers (N=197) were analysed previously by Palstra et al. (2007) and were added to this study. Samples (N=306) from two rivers further up the west coast of Newfoundland (Humber River and Western Arm Brook) were included for comparative purposes and in particular for the isolation-by-distance analysis (see below), bringing the grand total of samples to N=899. All scales were stored in dry paper envelopes from collection until molecular analyses. DNA was extracted from these samples and genetically characterized for 13 microsatellite marker loci, previously found to be selectively neutral, unlinked and free of technical artefacts in allele scoring for populations in this region (see Palstra et al. (2007, 2009) for details; locus SSsp1605 was omitted from the present study due to allele scoring difficulties).
Sample statistics
Basic descriptives for each sample and locus (allele frequencies, number of alleles, observed and expected heterozygosity) were obtained using FSTAT (version 2.9.3.2; Goudet, 1995). Genetic methods to estimate effective population size require selectively neutral marker loci and here we verified this assumption as follows. First, departures from Hardy–Weinberg equilibrium were assessed, for each locus and sample, by tests for departures of Fis from neutral expectations, as implemented in FSTAT (based on 5600 randomizations). The occurrence of genotyping errors (resulting from technical artefacts (null alleles) or DNA quality (large allele dropouts)) was checked using MICRO-CHECKER (van Oosterhout et al., 2004). Samples were rescored and amplification procedures (if possible) repeated, whenever irregularities were encountered. A total of 34 individuals were rescored for at least one locus (8% of total) and 25 samples were discarded due to failure to consistently amplify (2.8% of total).
Population structure
Genetic differentiation between all samples was estimated using pairwise FST (Weir and Cockerham, 1984) in GENETIX 4.04 (Belkhir et al., 2004). To assess the effects of age structure on temporal genetic stability (see Palstra and Ruzzante (2010)), these analyses were repeated on individual year cohorts, reconstructed based on age information of samples. We evaluated temporal stability of population structure by nesting temporal samples within rivers in a two-level hierarchical analysis of molecular variance using ARLEQUIN (Schneider et al., 2000). The analysis of molecular variance was also repeated on age cohorts nested within rivers. Population structure among rivers was further characterized by testing for isolation-by-distance relationships (Wright, 1943) on two different spatial scales: (1) a large spatial scale of nearly 250 km (encompassing all samples used in this study, including those from the Humber River and Western Arm Brook) and (2) a small spatial scale of <50 km (including only samples from the five rivers in the study system). Mantel tests (Mantel, 1967) were applied to evaluate the statistical significance of genetic–geographic distance associations, using the program IBD 1.52 (Bohonak, 2002).
We assessed the extent of spatial structure with the Bayesian clustering approach of STRUCTURE (Pritchard et al., 2000). This method calculates posterior probabilities for k clusters, assuming Hardy–Weinberg equilibrium conditions within clusters, while minimizing linkage disequilibrium. Runs were performed using a burnin length of 200 000 iterations followed by a Markov chain Monte-Carlo chain of equal length. These analyses were performed on the complete data set for the study system, including temporal replicates, and on a subset containing samples from three weakly differentiated populations in the centre of the system, those from Crabbes, Middle Barachois Brook and Robinsons (CMR). For both analyses, average posterior probabilities were calculated from five independent runs, for k ranging from 1 to 7.
Effective population size per river (N̂e(s))
We used two methods to estimate the contemporary effective population size of individual river populations. First, we applied the linkage disequilibrium method (Hill, 1981; England et al., 2006) to estimate (inbreeding) N̂e(LD). This method assumes that samples are derived from isolated populations, within which random mating occurs. We employed the program LDNe (Waples and Do, 2008) to estimate N̂e(LD) for each individual river sample. Alleles with an observed frequency P<0.02 were omitted from analyses, to maximize the trade-off between estimator bias and precision (Waples and Do, 2010). Confidence intervals associated with point estimates were calculated following equation (12) in Waples (2006).
Second, we used the temporally replicated samples in four out of five rivers to estimate (variance) N̂e(V) using the temporal method (Nei and Tajima, 1981; Waples, 1989). To reduce potential bias due to age structure (Waples and Yokota, 2007; Palstra and Ruzzante, 2008), we only used samples separated by at least 20 years, roughly four generations for Atlantic salmon in Newfoundland (Palstra et al., 2009). We used three different variations to estimate standardized temporal genetic variance (F) and N̂e(V): the moment-based estimators of Pollak (1983) and Jorde and Ryman (2007) and the pseudo-maximum likelihood method of Wang (2001). Again, rare alleles (P<0.02) were excluded from analyses. The last method (Wang and Whitlock, 2003) was also used to explore effects of gene flow on Ne estimation. Here, samples from other rivers in the study system were pooled to represent source populations.
The various contemporary Ne estimates assuming closed populations were combined into single composite estimates within each river, following the hierarchical method recommended by Waples and Do (2010). First, we estimated the harmonic mean of single sample (Ne(LD)) estimates obtained within rivers (that is, from temporally replicated collections), as well as the harmonic mean of within-river estimates of Ne(V) obtained with the three temporal methods mentioned above. Then, the harmonic mean of these two independent Ne estimates was calculated, to give a single estimate of N̂e(s) per river, that were subsequently used in one of the methods to estimate meta-Ne (see below).
Effective metapopulation size (meta-N̂e)
Effective metapopulation size (meta-N̂ewas calculated using three different approaches, for the weakly differentiated cluster of Robinsons, Crabbes and Middle Barachois (CMR), as well as for all five sampled rivers in the study system.
First, we estimated meta-Ne by simply adding the individual Ne estimates for each river (that is, ΣN̂e(s)). This provided a first reference point to investigate the effects of population subdivision on meta-Ne as estimated using the following two methods.
Second, a ‘top-down’ approach was applied where meta-Ne was estimated from pooled genetic data, considering individual river samples as derived from a single, though subdivided, population. We subsequently refer to these estimates as meta-N̂e(pooled). Samples were pooled only when they fell within one-generation length (∼5 years). We calculated the general magnitude of meta-N̂e(pooled) following Waples and Do (2010) as detailed for the per-river N̂e(s) estimates.
Third, we used the ‘bottom-up’ approach of Tufto and Hindar (2003). This approach combines estimates of effective size for individual populations (N̂e(s) calculated previously) and patterns of connectivity, to calculate meta-N̂e(T&H). Based on the population structure analyses, we defined a migration matrix based on a one-dimensional stepping-stone model (Wright, 1943). Ideally, assignment tests would have been used to estimate contemporary connectivity, but the weak differentiation among rivers prohibited this (see Faubet et al. (2007)). Instead, we specified a migration matrix with symmetric migration rates among adjacent rivers at the rate of m=0.05, reflecting average straying rates in this species (Stabell, 1984). Meta-N̂e(T&H) was estimated through minimizing the eigen-value of the resulting metapopulation matrix (Tufto and Hindar, 2003), employing the R library developed by J Tufto (available from http://www.math.ntnu.no/~jarlet/migration/).
To address our second objective of examining the issue of how gene flow asymmetries, differences in the effective size of individual populations and their interactions affect meta-Ne, we used simulations that explore the sensitivity of meta-N̂e(T&H) to uncertainty in N̂e(s). First, we separately varied input values of N̂e(s) for each river, from 50 to 150% of original estimates. Similarly, the consequences of asymmetrical migration for meta-N̂e(T&H) were explored under scenarios of positive (dispersal from large populations, mL, higher than dispersal from small populations, mS) and negative (lower dispersal from larger populations) density-dependent dispersal among rivers. In these simulations, Highlands was considered a small river, Robinsons and Flat Bay Brook were considered large, and Crabbes and Middle Barachois were considered intermediate (with dispersal intermediate between large and small). Simulations explored a wide range of magnitudes of gene flow (from m=0.005 to m=0.10) and imbalances therein between mL and mS (from mS=0.10mL to mS=10mL) to investigate the effects of gene flow asymmetry on meta-N̂e(T&H).
As we could not sample all rivers in the study system, we also explored the effects of non-sampled demes (Figure 1) by incorporating them into meta-N̂e(T&H). For this purpose, we approximated N̂e(s) of the seven non-sampled rivers (note that three of these waterways are very small and are unlikely to contain self-sustaining salmon populations) using a linear model including anadromous run size and drainage area (which were good predictors for N̂e(s) of rivers with data; r2=0.78, P=0.02). This analysis assumed a simple one-dimensional stepping-stone migration model with m=0.05.
Meta-Ne estimation and bias
The presence of bias due to incoming gene flow on N̂e(s), and population subdivision on meta-N̂e(pooled), was investigated through simulations in EASYPOP (Balloux, 2001). We simulated metapopulations consisting of five demes, characterized either by a one-dimensional stepping-stone model or by an island model. Deme sizes were set to Ne=500 (approximating the harmonic mean of N̂e(s)), characterized by random mating and equal sex ratios. We simulated 13 unlinked microsatellite marker loci, with μ=10−4 (cf. Fraser et al., 2007) and a maximum of 40 allelic states. Simulations were initiated with maximal gene diversity and run for 2000 generations (∼10 000 years in Atlantic salmon). Population connectivity was varied in these simulations (from m=0.0005 to m=0.20), with each scenario replicated 10 times. For each replicate run, we calculated N̂e(LD) using LDNe (Waples and Do, 2008) for each individual population sample, as well as for the data pooled from all demes (meta-N̂e(pooled)). Bias in the estimation of Ne due to incoming gene flow was evaluated by comparing N̂e(LD) with the expectation (Ne(s)=500). Individual N̂e(LD) estimates were then summed to obtain ΣN̂e(s) and were also combined with the migration matrix defined for each simulation, to estimate meta-N̂e(T&H). Under the idealized settings of these simulations, meta-N̂e(T&H) is expected to provide an unbiased estimate of meta-Ne (Tufto and Hindar, 2003). Hence, bias for the other two empirical meta-Ne estimates was assessed by comparison with meta-N̂e(T&H). However, bias was also investigated by assessing how meta-Ne estimates predicted the rate of loss of neutral genetic diversity in the entire metapopulation, as observed over the last 20 generations simulated.
Results
Basic descriptives
Genetic diversity among samples was moderate to high, with expected heterozygosities per locus ranging from 0.675 (Ssa-12) to 0.917 (Ssa-171), and heterozygosity per sample ranging from 0.749 (Middle Barachois-1973) to 0.835 (Middle Barachois-1998). The total number of alleles per sample ranged from 85 (Middle Barachois-1973) to 167 (Middle Barachois-1998), with a grand total of 268 alleles available for further analyses. 17 out of 247 tests (6.9%) for Hardy–Weinberg equilibrium were significant, a result expected by chance alone (at α=0.05, χ2-test, df=1, P=0.24). No deviation was consistent across loci or samples. MICRO-CHECKER indicated four potential occurrences of null alleles, again not consistent across samples, and we thus conclude that the allele frequencies used for subsequent analyses were unlikely to exhibit bias due to scoring or technical errors. See Supplementary Appendix A for a complete overview of gene marker characteristics.
Population structure
Genetic differentiation among samples, as measured by pairwise FST (Weir and Cockerham, 1984) was very weak to moderate. Among rivers, genetic differentiation ranged from FST=−0.0009 (Crabbes-2000 vs Robinsons-2001) to FST=0.069 (Middle Barachois-1982 vs Western Arm Brook-1982). Temporal stability among samples within rivers ranged from strong (FST=−0.0012, Robinsons-1982 vs Robinsons-2000) to weak (FST=0.0192, Middle Barachois-1973 vs Middle Barachois-1980). Significant isolation-by-distance patterns (Figure 2) were observed over the entire range sampled from the Highlands river in the south to the Western Arm Brook river in the north, as well as at the local scale (within 50 km) of the samples from the five rivers in the study system (see Table 1 and Figure 1).
An analysis of molecular variance on the samples from the study system suggested spatial and temporal components of genetic variance of similar magnitudes. When all five rivers were considered, 1.04% of genetic variance could be attributed to variance among rivers, with 0.33% attributed to variance among temporal samples within rivers (both variance components significant P<0.001). However, when these analyses were repeated on the subset of samples from Crabbes, Robinsons and Middle Barachois, the temporal variance within rivers (0.29%, P<0.001) was slightly larger than spatial variance among rivers (0.25%, P=0.059). In both cases, the percentage of variance explained was thus extremely low. A similar result was obtained when repeating the latter analyses on samples grouped into age cohorts (0.30% among cohorts (P<0.001) and 0.26% among rivers (P=0.026)).
These results are closely mirrored in the Bayesian clustering analyses of STRUCTURE. Analyses of samples from Crabbes, Robinsons and Middle Barachois suggest they form a single genetic cluster (highest posterior probability for K=1). Overall five rivers, the results from STRUCTURE analysis suggest K=5 is most likely, but these clusters did not clearly correspond to the individual rivers.
Together, the results of the population structure analyses suggest a dynamic system with weakly differentiated river populations, with connectivity best characterized by a one-dimensional stepping-stone model of gene flow, indicated by the isolation-by-distance pattern observed (Figure 2). These results, therefore, suggest the use of a stepping-stone model of connectivity in subsequent analyses is appropriate.
Effective population size (N̂e(s)) per river
Based on single samples, estimates of Ne(LD) ranged from 188 (Middle Barachois-1975) to ∞ (in several samples) (Table 2). Ne(LD) estimates also displayed considerable variation over time within the same river. Harmonic mean N̂e(LD) per river ranged from 204 to ∞. Temporal estimates of Ne(v) (Table 3) were possible for four rivers and ranged from 38 (Flat Bay Brook) to 1110 (Middle Barachois). The moment-based estimator of Jorde and Ryman (2007) and the pseudo-likelihood approach (Wang, 2001) yielded results that were qualitatively consistent (Table 3). For each river, estimates of Ne(v) using the temporal approach of Wang and Whitlock (2003) including gene flow were smaller than those assuming closed populations (see Supplementary Appendix B), with gene flow estimates comparable to dispersal rates typically observed in Atlantic salmon (Stabell, 1984). Combining estimates of the temporal methods assuming closed populations resulted in harmonic mean Ne(v) estimates per river ranging from 54 (Flat Bay) to 849 (Middle Barachois) (Table 3).
Although the different estimators apply to slightly different time scales and may thus not be strictly comparable, they all provide independent indications of the general magnitude of contemporary Ne of the salmon populations in these rivers. Thus, composite N̂e(s) estimates per river were 204 (Highlands), 433 (Crabbes), 496 (Middle Barachois), 1347 (Flat Bay) and 1646 (Robinsons).
Effective metapopulation size (meta-Ne)
ΣN̂e(s)
First, summing the harmonic mean estimates for individual rivers gave ΣN̂e(s)=2575 for CMR and ΣN̂e(s)=4126 for all five study rivers. Logically, this estimate is larger when all five rivers are included than when only the three weakly differentiated central rivers (CMR) are considered.
Meta-N̂e(pooled)
Second, for the set of three weakly differentiated rivers (CMR), the linkage disequilibrium method gave meta-N̂e(pooled) ranging from 706 (samples 1980–1982 pooled) to ∞ (samples 1998–2001 pooled) (Table 2). We used these two pooled samples for the temporal methods, which gave meta-N̂e(pooled) ranging from 1071 to 2234 (Table 3). Second, we estimated ‘total’ metapopulation effective size by pooling, where possible, genetic data from all rivers. Meta-N̂e(pooled) estimation for all five rivers, using the linkage disequilibrium method, was possible only for the combined samples from 1998 to 2001, giving meta-N̂e(pooled)=729. Using this combined sample (1998–2001) in the temporal approach with a combined sample from CMR-1980 as t0, gave meta-N̂e(pooled)=443. Combining and weighting these estimates (cf. Waples and Do, 2010) gave meta-N̂e(pooled)=2097 for CMR and meta-N̂e(pooled)=665 for all five rivers. Hence, with this ‘pooling rivers’ approach, meta-N̂e(pooled) is larger for the three central rivers (CMR) than it is for all five rivers.
Meta-N̂e(T&H) and comparisons
Third, the harmonic mean per-river N̂e(s) values were used for the calculation of meta-N̂e(T&H). Assuming a one-dimensional stepping-stone model with symmetrical gene flow among neighbouring rivers, meta-N̂e(T&H)=1835 for CMR and meta-N̂e(T&H)=2418 for all five rivers. Thus, meta-N̂e(pooled) and meta-N̂e(T&H) generally gave estimates of meta-Ne that were smaller than ΣN̂e(s), but the extent of this difference appears related to the extent of genetic differentiation among populations. In fact, meta-N̂e(pooled) was similar to meta-N̂e(T&H) for CMR, whereas overall five rivers, where population structure was stronger meta-N̂e(pooled) was only 28% of meta-N̂e(T&H).
Sensitivity analysis
Simulation was used to evaluate which individual deme Ne estimates has the strongest influence on meta-Ne. First, for the CMR cluster, the estimate for Crabbes (having the smallest harmonic mean N̂e(s)) exerts the largest influence on meta-N̂e(T&H) (Figure 3, top). Conversely, over all five rivers, uncertainty about N̂e(s) of the two outermost rivers (Highlands and Flat Bay Brook) has the strongest influence on meta-N̂e(T&H) (Figure 3, bottom). Second, for the CMR group, the difference between meta-N̂e(T&H) and meta-N̂e(pooled) can be explained by relatively small bias in any of the individual N̂e(s) values. Conversely, over all five rivers (upward) bias in any of the Ne(s) estimates appears unlikely to be the explanation for the reduction in meta-N̂e(pooled) relative to meta-N̂e(T&H).
Relative sensitivity of meta-N̂e(T&H) to uncertainty in deme Ne estimates, quantified by varying model input values of individual deme Ne estimates (from 50 to 150% of N̂e(s)) for a weakly differentiated subset of rivers (top, d=3 consisting of Crabbes, Robinsons and Middle Barachois Brook) and for all five rivers (bottom, d=5). Also given are the relative magnitudes of meta-N̂e(pooled) to meta-N̂e(T&H) for both sets of samples (solid grey lines).
Simulations of the effects of gene flow asymmetry on meta-N̂e(T&H) (Figure 4, left) suggest that positive density-dependent (higher dispersal from larger populations) dispersal tends to increase meta-N̂e(T&H), whereas negative density-dependent dispersal reduces meta-N̂e(T&H). Importantly, meta-N̂e(T&H) is affected primarily not by the magnitude of gene flow, but by the degree of gene flow asymmetry among populations. These trends are similar when smaller input values of N̂e(s) are used (Figure 4, right). Therefore, over all five rivers, concordance between meta-N̂e(pooled) and meta-N̂e(T&H) requires strong negative density-dependent dispersal (for example, mS=10mL), smaller N̂e(s) values as input (that is, the original estimates are biased upwardly) or a combination of these two.
Sensitivity of meta-N̂e(T&H) to gene flow asymmetry. Shown are results for various gene flow scenarios, where dispersal from small populations (mS) is varied relative to dispersal from large population (mL), over the range of gene flow rates commonly observed in Atlantic salmon. mS<1.0 indicates positive density-dependent dispersal, mS>1.0 indicates negative density-dependent dispersal and mS=mL indicates gene flow symmetry among neighbouring demes. Results of simulations are given for scenarios based on original N̂e(s) values (left panel) and scenarios using 50% of N̂e(s) as input values (right panel).
Finally, an extension of these analyses to all rivers in the study system gave meta-N̂e(T&H)=1913. Although harmonic mean N̂e(s) was smaller than for the situation where only five rivers were analysed, the number of demes also increased, and the resulting meta-N̂e(T&H) remains much larger than meta-N̂e(pooled) for the five rivers. Upward bias in meta-N̂e(T&H) due to the omission of some (small) rivers in its original estimation may thus be unlikely to be an explanation for its large magnitude relative to meta-N̂e(pooled).
Simulations of meta-Ne
Simulations using EASYPOP suggest individual population Ne(LD) estimates can be biased upwardly, but only when incoming gene flow is moderate to high (in simulations with Ne(s)=500 and m>0.05 or FST<0.01, Figure 5, top panels). Thus, the discrepancy observed between the two empirical meta-Ne estimation methods for all five rivers might not be due to inflation in meta-N̂e(T&H) caused by a general upward bias in the underlying individual Ne(s) estimates.
Evaluation of potential bias in meta-Ne estimates. Given are comparisons between deme Ne(s) estimates and the expected value of Ne(s)=500 used in simulations (bias-N̂e(s) expressed as the ratio N̂e(s)/500), for a range of genetic differentiation among demes in the island model (top left) and stepping-stone model (top right). Also given are comparisons of meta-Ne estimates based on pooled genetic data (x meta-N̂e(pooled)), on the model of Tufto and Hindar (2003) (• meta-N̂e(T&H)) and on the sum of individual deme estimates (○ ΣN̂e(s)) for a range of values of genetic differentiation among demes, for the island model (bottom left) and stepping-stone model (bottom right). Simulations performed in EASYPOP and empirical Ne estimates calculated using software LDNe.
Second, population subdivision may introduce downward bias into meta-N̂e(pooled) estimates, but not into estimates of meta-N̂e(T&H), since the latter method explicitly accounts for structure. Results are similar for the stepping-stone and island models and will thus be presented as one (Figure 5, bottom panels). As genetic differentiation among demes increases due to reduced gene flow, meta-N̂e(pooled) estimates decrease exponentially, compared with meta-N̂e(T&H) estimates. For both models of population structure (stepping-stone or island), ΣN̂e(s) were consistently larger than meta-N̂e(T&H) (paired t-test, two-tailed, P=0.0014) but this difference appears unaffected by the extent of genetic differentiation.
Finally, the rates of loss of neutral genetic diversity observed in the simulated metapopulations generally suggest these are predicted much more closely by meta-N̂e(T&H) than by meta-N̂e(pooled). These simulations therefore indicate that population subdivision (spatial structure) introduces downward bias into empirical estimation of meta-Ne when this structure is not explicitly accounted for.
Discussion
In this study, we have examined the factors that affect the estimation of the effective size of a metapopulation (meta-Ne) using three approaches: simply adding the estimates of individual population effective sizes (ΣN̂e(s)), pooling all data before estimation (meta-N̂e(pooled)) and using the methodology developed by Tufto and Hindar (2003), which requires knowledge of Ne(s) of individual demes and their connectivity within the metapopulation system (meta-N̂e(T&H)). We have examined the performances of these three methods under conditions of high and low gene flow, gene flow asymmetries encompassing positive and negative density-dependent dispersal, as well as the uncertainties in the estimates of individual Ne(s). Our main findings can be summarized as follows. Under very weak genetic differentiation (that is, FST<0.005) as exemplified by the three weakly differentiated central populations in our Atlantic salmon system (CMR rivers), estimates of meta-N̂e(pooled) and meta-N̂e(T&H) were similar to each other, but slightly smaller than the sum of individual population, or deme, effective sizes (ΣN̂e(s)). Conversely, under stronger genetic differentiation among demes, as observed for all five rivers, meta-N̂e(pooled) was 27% of meta-N̂e(T&H) and 16% of ΣN̂e(s). These observations, therefore, support the general expectation that population subdivision reduces meta-Ne (cf. Whitlock, 2004). However, the observed discrepancy between meta-N̂e(pooled) and the meta-N̂e(T&H) methods requires an explanation. We, thus, examined the roles of strong negative density-dependent dispersal among populations, a general upward bias in individual river N̂e(s) estimates, and their interactions. Our findings are consistent with the notion that both asymmetrical gene flow and unequal population sizes can reduce meta-Ne (Whitlock and Barton, 1997; Whitlock, 2004). However, the reductions in meta-Ne estimates are minimal under conditions of positive density-dependent dispersal (higher dispersal from larger populations). We discuss in detail below, the possibilities that (1) individual deme Ne(s) estimates may have been upwardly biased; (2) meta-N̂e(pooled) estimates may have been downwardly biased; (3) our results are affected by non-sampled populations and/or (4) assumptions implicit to the model of Tufto and Hindar (2003) may be violated in Atlantic salmon.
Upward bias in individual Ne estimates
Role of age structure and gene flow
First, individual Ne(s) estimates may have been biased upwardly. We attempted to minimize bias due to age structure in the temporal methods (Waples and Yokota, 2007; Palstra and Ruzzante, 2008) by only considering longer time spans. As we have done before (Palstra et al., 2009), we pooled data from several consecutive years to attain allele frequency estimates more closely reflecting those of the entire population. Regardless, ignored age structure is most likely to introduce a downward bias into Ne(s) estimates (Waples and Yokota, 2007). Also, we generally ignored gene flow, which may be particularly relevant for the three central rivers exhibiting weak spatial genetic differentiation (Crabbes, Middle Barachois and Robinsons). The linkage disequilibrium method, however, has been reported to be fairly robust to gene flow (Waples and Do, 2010) and this is supported by our simulations (Figure 5, bottom panels). Conversely, estimates from the temporal method including gene flow (Wang and Whitlock, 2003) were all lower than those ignoring gene flow (Supplementary Appendix B). Although the biological relevance of this temporal approach remains uncertain (Hoffman et al., 2004; Fraser et al., 2007; Palstra and Ruzzante, 2008), these conflicting results suggest we cannot rule out the possibility of upward bias in N̂e(s) and meta-N̂e(T&H) caused by incoming gene flow. However, the fact that meta-N̂e(pooled) and meta-N̂e(T&H) are very similar under conditions of high gene flow (that is, the CMR cluster in this study) suggests a general upward bias in N̂e(s) might be an unlikely explanation for the patterns we observed.
Population subdivision and additional linkage disequilibrium
Our EASYPOP simulations suggest that population subdivision itself may introduce additional signals of linkage disequilibrium or inbreeding, akin to Wahlund effects (Wahlund, 1928), thereby introducing downward bias into the meta-N̂e(pooled) estimation. Further support for this downward bias comes from the observed temporal rates of loss of genetic variance in the simulated metapopulations, which always corresponded much more closely to meta-N̂e(T&H) than to meta-N̂e(pooled). This downward bias in meta-N̂e(pooled) may thus provide one explanation for the observed discrepancy between the two empirical meta-Ne estimates under conditions of low gene flow (that is, all five river populations in this study).
In addition to the linkage effect above, meta-Ne estimates based on pooled genetic data may have been biased (downwardly) due to unbalanced and incomplete sampling of rivers. Sampling limitations resulted in estimates of N̂e(pooled) that were sometimes based on a subset of the sampled rivers, which may have introduced downward bias, particularly for the temporal methods. Yet, it should have affected single sample estimates less than two-sample temporal estimates, but the two methods gave estimates of meta-Ne that were of similar magnitude. Thus, we suspect additional linkage brought on by the pooling of data from genetically divergent populations may be a more important effect than incomplete sampling.
Third, the present study only sampled five of nine salmon rivers in the Bay of St George area (which also has three other streams likely too small to hold their own Atlantic salmon populations; J Brian Dempson, personal communication). However, meta-N̂e(T&H) estimates, with or without the unsampled populations, remained large relative to meta-N̂e(pooled). Although future studies should include these non-sampled rivers, we suspect they are unlikely to account for the small value of meta-N̂e(pooled) relative to meta-Ne(T&H).
Finally, the assumptions in the model of Tufto and Hindar (2003) need to be reviewed. Application of this model first requires knowledge of migration rates. We necessarily had to adopt an approximation approach, as actual migration rates among demes were unknown due to the very weak genetic differentiation observed. However, the most critical assumption of this model is probably that the effective size of individual demes is constant over time, hence leading to the convergence of meta-Ne onto the eigen-value in this model. Yet, fluctuations in population size over time represent one of the main demographic factors reducing Ne below census population size (Vucetich et al., 1997; Waples, 2002). In the extreme case, local population extinction can strongly reduce the effective size of a metapopulation (Whitlock and Barton, 1997). These considerations may limit the applicability of the model of Tufto and Hindar (2003) to natural populations, although monitoring of populations over time should reveal such extreme population fluctuations. Violation of the assumption of constant subpopulation sizes awaits further assessment, for example with simulations, to better understand the empirical utility of this estimator method.
Factors affecting the effective size of metapopulations
This study adds some new insights into the factors shaping the effective size of metapopulations in Atlantic salmon. First, metapopulation effective size in this species appears to be (much) smaller than the sum of the deme sizes (ΣN̂e(s)) and this reduction increases with increasing genetic differentiation among demes. Results presented here also suggest asymmetrical migration may be an important factor, or requirement for, observations of reduced meta-Ne. Kuparinen et al. (2010) reported that the effectively smallest population had the strongest effect on meta-Ne, mainly because it also had the highest emigration rate, hence corresponding to a scenario of negative density-dependent dispersal. Our simulations confirm that negative density-dependent dispersal patterns can strongly reduce meta-Ne, more so than positive density-dependent dispersal can. It is not known, however, how prevalent such connectivity patterns are in the wild. Based on demography and neutral conditions, one would expect positive density-dependent dispersal, and so far this pattern is supported by empirical genetic analyses (for example, Hansen et al., 2007, but see Palstra et al. (2007)). Although our results are most easily explained by a combination of negative density-dependent dispersal and a general upward bias in Ne(s) estimates (Figure 4, left), these two requirements may be somewhat mutually exclusive. For example, Ne(s) estimates for presumably larger populations are unlikely to be biased upwardly due to incoming gene flow from relatively smaller populations.
Genetic differentiation itself can also introduce downward bias into the empirical estimation of Ne for a subdivided population, when this subdivision is ignored in analyses. The results from simulations (Figure 5) were also observed in a meta-analysis of empirical data (Supplementary Appendix C). Hence, this bias appears present under empirical settings, and will increase with increasing genetic differentiation among demes. Only when connectivity among demes is very high (simulations: FST<0.005; Figure 5, bottom panels) might meta-N̂e(pooled) provide relatively unbiased indications of meta-Ne. Downward bias already becomes relevant under limited genetic differentiation (FST∼0.01, Figure 5, top panels). This inference thus argues against the general use of meta-N̂e(pooled), thereby unfortunately removing a reference point for assessing the performance of other meta-Ne estimators. The development of additional empirical approaches to estimate meta-Ne thus seems both timely and desirable.
Overall, this study provides some support for the suggestion (Palstra and Ruzzante, 2008) that estimates of the N̂e/N ratio for widespread marine organisms may be downwardly biased by population subdivision. Interestingly, Hoarau et al. (2005) reported that their low N̂e/N ratio for North Sea plaice could be explained by inbreeding. This inference may essentially be correct, with the distinction that population subdivision (that is, inbreeding), may have introduced a downward bias into N̂e, in addition to inbreeding effects reducing Ne through biological processes. This highlights the general need to make a distinction between factors affecting the behaviour of the population parameter Ne and those affecting its empirical estimation.
Finally, this study has some implications for the perception of the regulation of genetic variance required to maintain evolutionary potential. In salmonid fish and other organisms, the magnitude of estimates of contemporary Ne for many individual populations generally suggests that these may be too small to maintain long-term adaptability (Palstra and Ruzzante, 2008). Conversely, contemporary meta-Ne estimates presented here are much closer to benchmark magnitudes of Ne (Franklin and Frankham, 1998; Lynch and Lande 1998; Willi et al., 2006) and also to long-term Ne estimates (for example, Fraser et al., 2007; Palstra et al., 2007). The genetic variance maintained in a metapopulation is shaped by long-term processes, whereas the effective size of a local population may be primarily sensitive to evolutionary processes occurring on the scale of one or a few generations (Waples, 2010). Hence, low contemporary Ne estimates for local populations do not necessarily imply low adaptability when the genetic variance is principally maintained at the higher hierarchical level of a metapopulation. Continuing species declines and habitat fragmentation will, however, affect the relevance of this conclusion.
References
Balloux F (2001). EASYPOP (Version 1.7): a computer program for population genetics simulations. J Hered 92: 301–302.
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, Logiciel sous Windows TM pour la génétique des populations 1996–2004. 04. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II: Montpellier (France).
Bohonak AJ (2002). IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 93: 153–154.
Caballero A (1994). Developments in the prediction of effective population size. Heredity 73: 657–679.
Caballero A, Hill WG (1992). Effective size of nonrandom mating populations. Genetics 130: 909–916.
Chaput G, Dempson JB, Caron F, Jones R, Gibson J (2006). A synthesis of life history characteristics and stock grouping of Atlantic salmon (Salmo salar L) in eastern Canada. Canadian Science Advisory Secretariat Research Document 2006/015.
Consuegra S, Verspoor E, Knox D, Garciá De Leániz C (2005). Asymmetric gene flow and the evolutionary maintenance of genetic diversity in small, peripheral Atlantic salmon populations. Conserv Genet 6: 823–842.
Dempson JB, Clarke G (2001). Status of the Atlantic salmon (Salmo salar) at Highlands River, Bay St George, SFA 13, Newfoundland, 2000. Canadian Science Advisory Secretariat Research Document 2001/028.
Dionne M, Caron F, Dodson JJ, Bernatchez L (2008). Landscape genetics and hierarchical genetic structure in Atlantic salmon: the interaction of gene flow and local adaptation. Mol Ecol 17: 2382–2396.
Engen S, Lande R, Saether BE (2005). Effective size of a fluctuating age-structured population. Genetics 170: 941–954.
England PR, Cornuet JM, Berthier P, Tallmon DA, Luikart G (2006). Estimating effective population size from linkage disequilibrium: severe bias in small samples. Conserv Genet 7: 303–308.
Faubet P, Waples RS, Gaggiotti OE (2007). Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16: 1149–1166.
Frankham R (1995). Effective population size adult population size ratios in wildlife—a review. Genet Rese 66: 95–107.
Franklin IR, Frankham R (1998). How large must populations be to retain evolutionary potential? Anim Conserv 1: 69–73.
Fraser DJ, Hansen MM, Østergaard S, Tessier N, Legault M, Bernatchez L (2007). Comparative estimation of effective population sizes and temporal gene flow in two contrasting population systems. Mol Ecol 16: 3866–3889.
Gaggiotti OE, Vetter RD (1999). Effect of life history strategy, environmental variability, and overexploitation on the genetic diversity of pelagic fish populations. Can J Fish Aquat Sci 56: 1376–1388.
Goudet J (1995). FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86: 485–486.
Hansen MM, Skaala O, Jensen LF, Bekkevold D, Mensberg KLD (2007). Gene flow, effective population size and selection at major histocompatibility complex genes: brown trout in the Hardanger Fjord, Norway. Mol Ecol 16: 1413–1425.
Hare M, Nunney L, Schwartz MK, Ruzzante DE, Burford M, Waples RS, Ruegg K, Palstra FP (2011). Understanding and estimating effective population size for practical application in marine species management. Conserv Biol (doi:10.1111/j.1523-1739.2010.01637.x).
Hendry AP, Castric V, Kinnison MT, Quinn TP (2004). The evolution of philopatry and dispersal. Homing versus straying in salmonids. In: Hendry AP, Stearns SC (eds). Evolution Illuminated. Salmon and Their Relatives. Oxford University Press: New York, pp 52–91.
Hill WG (1981). Estimation of effective population size from data on linkage disequilibrium. Genet Res 38: 209–216.
Hindar K, Tufto J, Sættem LM, Balstad T (2004). Conservation of genetic variation in harvested salmon populations. ICES J Mar Sci 61: 1389–1397.
Hoarau G, Boon E, Jongma DN, Ferber S, Palsson J, Van Der Veer HW, Rijnsdorp AD, Stam WT, Olsen JL (2005). Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleuronectes platessa L.). Proc R Soc B Biol Sci 272: 497–503.
Hoffman EA, Schueler FW, Blouin MS (2004). Effective population sizes and temporal stability of genetic structure in Rana pipiens, the northern leopard frog. Evolution 58: 2536–2545.
Jorde PE, Ryman N (1995). Temporal allele frequency change and estimation of effective size in populations with overlapping generations. Genetics 139: 1077–1090.
Jorde PE, Ryman N (2007). Unbiased estimator for genetic drift and effective population size. Genetics 177: 927–935.
King TL, Kalinowski ST, Schill WB, Spidle AP, Lubinski BA (2001). Population structure of Atlantic salmon (Salmo salar L.): a range-wide perspective from microsatellite DNA variation. Mol Ecol 10: 807–821.
Kuparinen A, Tufto J, Consuegra S, Hindar K, Merilä J, Garciá De Leániz C (2010). Effective size of an Atlantic salmon (Salmo salar L.) metapopulation in Northern Spain. Conserv Genet 11: 1559–1565.
Levins R (1969). Some demographic and genetic consequences of environmental heterogeneity for biological control. Bull Entomol Soc Am 15: 237–240.
Li CC (1955). Population Genetics. University of Chicago Press: Chicago, IL.
Lynch M, Lande R (1998). The critical effective size for a genetically secure population. Anim Conserv 1: 70–72.
Mantel N (1967). The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220.
Morrissey MB, de Kerckhove DT (2009). The maintenance of genetic variation due to asymmetric gene flow in dendritic metapopulations. Am Nat 174: 875–889.
Mullins CC, Caines D (2000). Status of the Atlantic salmon (Salmo salar) stock of Humber River, Newfoundland, 1999. Canadian Stock Assessment Secretariat Research Document 2000/037.
Mullins CC, Caines D, Lowe SL (2000). Status of the Atlantic salmon (Salmo salar) stock of Harry's River/Pinchgut Brook, Newfoundland, 2000. Canadian Science Advisory Secretariat Research Document 2001/033.
Nei M, Tajima F (1981). Genetic drift and estimation of effective population size. Genetics 98: 625–640.
Nunney L (1999). The effective size of a hierarchically structured population. Evolution 53: 1–10.
Nunney L, Elam DR (1994). Estimating the effective population size of conserved populations. Conserv Biol 8: 175–184.
O’Connell MF, Ash EGM (1993). Smolt size in relation to age at 1st maturity of Atlantic Salmon (Salmo salar)—the role of lacustrine habitat. J Fish Biol 42: 551–569.
O’Connell MF, Dempson JB, Chaput G (2006). Aspects of the life history, biology and population dynamics of Atlantic salmon (Salmo salar L.) in eastern Canada. Canadian Science Advisory Secretariat Research Document 2006/014.
Østergaard S, Hansen MM, Loeschcke V, Nielsen EE (2003). Long-term temporal changes of genetic composition in brown trout (Salmo trutta L.) populations inhabiting an unstable environment. Mol Ecol 12: 3123–3135.
Palstra FP, Ruzzante DE (2008). Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol Ecol 17: 3428–3447.
Palstra FP, Ruzzante DE (2010). A temporal perspective on population structure and gene flow in Atlantic salmon (Salmo salar) in Newfoundland, Canada. Can J Fish Aquat Sci 67: 225–242.
Palstra FP, O’Connell MF, Ruzzante DE (2007). Population structure and gene flow reversals in Atlantic salmon (Salmo salar) over contemporary and long-term temporal scales: effects of population size and life history. Mol Ecol 16: 4504–4522.
Palstra FP, O’Connell MF, Ruzzante DE (2009). Age structure, changing demography and effective population size in Atlantic salmon (Salmo salar). Genetics 182: 1233–1249.
Pollak E (1983). A new method for estimating the effective population size from allele frequency changes. Genetics 104: 531–548.
Porter TR, Clarke G, Murray J (2002). Status of Atlantic salmon (Salmo salar L.) populations in Crabbes and Robinsons Rivers, and Middle Barachois, Fischells and Flat Bay Brooks, Newfoundland, 2001. Canadian Science Advisory Secretariat Research Document 2002/029.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Schneider S, Kueffer J-M, Roessli D, Excoffier L (2000). Arlequin, Version 2. A Software for Population Genetics Data Analysis. Genetics and Biometry Laboratory, University of Geneva: Geneva.
Schwartz MK, Luikart G, Waples RS (2007). Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22: 25–33.
Stabell OB (1984). Homing and olfaction in salmonids—a critical review with Special reference to the Atlantic salmon. Biol Rev Camb Philos Soc 59: 333–388.
Sugg DW, Chesser RK, Dobson FS, Hoogland JL (1996). Population genetics meets behavioral ecology. Trends Ecol Evol 11: 338–340.
Tufto J, Hindar K (2003). Effective size in management and conservation of subdivided populations. J Theor Biol 222: 273–281.
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538.
Verspoor E, Stradmeyer L, Nielsen JL (2008). The Atlantic Salmon: Genetics, Conservation and Management. Blackwell Publishing: Oxford.
Vucetich JA, Waite TA, Nunney L (1997). Fluctuating population size and the ratio of effective to census population size. Evolution 51: 2017–2021.
Wahlund S (1928). The combination of populations and the appearance of correlation examined from the standpoint of the study of heredity. Hereditas 11: 65–106.
Wang JL (1996). Inbreeding and variance effective size for nonrandom mating populations. Evolution 50: 1786–1794.
Wang JL (2001). A pseudo-likelihood method for estimating effective population size from temporally spaced samples. Genet Res 78: 243–257.
Wang JL, Caballero A (1999). Developments in predicting the effective size of subdivided populations. Heredity 82: 212–226.
Wang JL, Whitlock MC (2003). Estimating effective population size and migration rates from genetic samples over space and time. Genetics 163: 429–446.
Waples RS (1989). A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics 121: 379–391.
Waples RS (2002). Effective size of fluctuating salmon populations. Genetics 161: 783–791.
Waples RS (2006). A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7: 167–184.
Waples RS (2010). Spatial-temporal stratifications in natural populations and how they affect understanding and estimation of effective population size. Mol Ecol Resour 10: 785–796.
Waples RS, Do C (2008). LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8: 753–756.
Waples RS, Do C (2010). Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3: 244–262.
Waples RS, Yokota M (2007). Temporal estimates of effective population size in species with overlapping generations. Genetics 175: 219–233.
Wares JP, Pringle JM (2008). Drift by drift: effective population size is limited by advection. BMC Evol Biol 8: 235.
Weir BS, Cockerham CC (1984). Estimating F-Statistics for the analysis of population structure. Evolution 38: 1358–1370.
Whitlock MC (2004). Selection and drift in metapopulations. In: Hanski I, Gaggiotti OE (eds). Ecology, Genetics and Evolution of Metapopulations. Elsevier Academic Press: San Diego, CA, pp 153–175.
Whitlock MC, Barton NH (1997). The effective size of a subdivided population. Genetics 146: 427–441.
Willi Y, Van Buskirk J, Hoffmann AA (2006). Limits to the adaptive potential of small populations. Annu Rev Ecol Evol Syst 37: 433–458.
Wright S (1931). Evolution in Mendelian populations. Genetics 16: 97–159.
Wright S (1938). Size of a population and breeding structure in relation to evolution. Science 87: 430–431.
Wright S (1943). Isolation by distance. Genetics 28: 114–138.
Wright S (1969). Evolution and the Genetics of Populations, Vol. 2. The Theory of Gene Frequencies. University of Chicago Press: Chicago, IL.
Acknowledgements
Don Caines, J Brian Dempson, Geoff Veinott and Arthur Walsh (Fisheries and Oceans Canada) generously provided archived scale samples and background information on the study system. Abby van der Jagt assisted with the molecular genetic analyses. Robin Waples is thanked for providing feedback on a number of queries and for sharing an unpublished manuscript. Four anonymous reviewers provided many constructive comments on earlier versions of this manuscript. This work was supported by an NSERC Discovery grant to DER.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Palstra, F., Ruzzante, D. Demographic and genetic factors shaping contemporary metapopulation effective size and its empirical estimation in salmonid fish. Heredity 107, 444–455 (2011). https://doi.org/10.1038/hdy.2011.31
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2011.31
Keywords
This article is cited by
-
Contrasts in riverscape patterns of intraspecific genetic variation in a diverse Neotropical fish community of high conservation value
Heredity (2023)
-
Assessing conservation risks to populations of an anadromous Arctic salmonid, the northern Dolly Varden (Salvelinus malma malma), via estimates of effective and census population sizes and approximate Bayesian computation
Conservation Genetics (2017)
-
Origins and genetic diversity among Atlantic salmon recolonizing upstream areas of a large South European river following restoration of connectivity and stocking
Conservation Genetics (2014)