Abstract
Although similar to any other organism, prokaryotes can transfer genes vertically from mother cell to daughter cell, they can also exchange certain genes horizontally. Genes can move within and between genomes at fast rates because of mobile genetic elements (MGEs). Although mobile elements are fundamentally self-interested entities, and thus replicate for their own gain, they frequently carry genes beneficial for their hosts and/or the neighbours of their hosts. Many genes that are carried by mobile elements code for traits that are expressed outside of the cell. Such traits are involved in bacterial sociality, such as the production of public goods, which benefit a cell's neighbours, or the production of bacteriocins, which harm a cell's neighbours. In this study we review the patterns that are emerging in the types of genes carried by mobile elements, and discuss the evolutionary and ecological conditions under which mobile elements evolve to carry their peculiar mix of parasitic, beneficial and cooperative genes.
Similar content being viewed by others
Introduction
Although in all organisms genes are transferred vertically from parent to offspring, many microorganisms can also transfer genes horizontally, independently of reproduction events, through processes that are known collectively as horizontal gene transfer (HGT). Ever since Joshua Lederberg's pioneering work, it has been known that bacteria can exchange genes horizontally (Lederberg and Tatum, 1946; Zinder and Lederberg, 1952). However, it is only recently that both the significance and the scale of HGT has become apparent (Ochman et al., 2000; Bushman, 2002; Kurland et al., 2003; Bordenstein and Reznikoff, 2005; Sorensen et al., 2005; Thomas and Nielsen, 2005) and has even been described by one party as ‘biology's next revolution’ (Goldenfeld and Woese, 2007).
Prokaryotic genomes are now known to vary in the rates of gene gain and loss. Most prokaryotes are highly dynamic with high rates of gene gain and loss, whereas some genomes, typically associated with obligatory endosymbiosis, have stable or shrinking genomes. Within genomes, genes differ in their propensity to be mobile. In addition to the core genome (∼2000 genes in Escherichia coli that are present in the first 20 sequenced strains), non-core genes contribute significantly to the overall diversity of gene repertoires in a species, which together with the core are known as the pan-genome. In a study of E. coli, for example, these non-core genes made up 90% of the pan-genome when 20 strains were put together (Touchon et al., 2009). Despite the widespread attention that HGT has received in recent years (Delsuc et al., 2005; Frost et al., 2005; Gevers et al., 2005; Sorensen et al., 2005; Goldenfeld and Woese, 2007), the adaptive significance of horizontally transferred genes remains to be fully addressed.
There are increasing hints of important separations of functions between the vertically transmitted core genome, which encodes fundamental cellular processes, and the horizontally transmissible accessory genome, which encodes for a variety of secondary metabolites conferring resistance to specific toxins or antibiotics or the ability to exploit a specific niche (Hacker and Carniel, 2001; Norman et al., 2009). The accessory genome contains recently acquired functions, mobile genetic elements (MGEs), non-expressed genes and genes under particular modes of selection such as diversifying selection, frequency-dependent selection and periodic selection. Despite a large and growing body of research into the molecular mechanisms of HGT, the ecological and evolutionary forces that drive these basic divisions of mobility and function are poorly understood (see, for example, Slater et al., 2008).
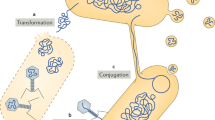
Horizontal transfer exists largely because some MGEs can move or be mobilized between microbial cells and carry other traits than the ones strictly essential for their replication (Frost et al., 2005). They encompass a wide set of different elements, such as transposable elements, plasmids and bacteriophages. In prokaryotes, mobile genes can be transmitted horizontally in three main ways: transformation (in which naked DNA is taken up from the local environment; Chen and Dubnau, 2004), transduction (transfer of DNA through a bacteriophage vector) or by conjugation (transfer of DNA through cell–cell contact, most often mediated by plasmids; Cascales and Christie, 2003; Thomas and Nielsen, 2005; de la Cruz et al., 2009). Horizontally transferred genes confer a variety of beneficial and negative effects on their bacterial hosts.
The costs of mobile elements differ significantly with the element. Bacteriophages are commonly divided into virulent and temperate. Upon successful invasion, virulent phages kill the cell to release their progeny (although filamentous phages are able to export progeny without cell lysis). Temperate phages may exert an effect similar to virulent phages, but they may also integrate in the chromosome and replicate with the bacteria (as a quiescent prophage), and some can reside in the cell as plasmid prophages. In this dormant state, the microbial cell may even express useful traits carried by the phages, yet so long as the phage remains functional, the possibility remains that the phage will exit from the quiescent state, kill the host and transmit horizontally.
The introduction of other mobile elements, such as plasmids, integrative conjugative elements, mobilizable islands or transposable elements, does not pose the same life-or-death dilemma. However, these elements are still costly (Diaz Ricci and Hernandez, 2000). Element replication requires the synthesis of proteins, RNA and DNA, which incur in a fitness cost. This cost can be low in general and easily surmounted by other adaptive or addictive traits. Mobile elements often integrate in chromosomes, thereby potentially disrupting important functions (Lerat and Ochman, 2004). Transmission often involves the creation of structures, such as conjugation pili, that can be costly. These structures can themselves facilitate the invasion of phages (Rasched and Oberer, 1986). In the case of elements carried by virulent phages, transmission results in host death. Mobile elements can also be costly because of the genes they carry to compete with other genetic elements. For example, F-like plasmids use exclusion proteins to prevent superinfection by other closely related plasmids that are very highly expressed (Achtman, 1975).
Mobile elements may also carry genes coding for adaptive traits. They have thus been described as ‘agents of open source evolution’ (Frost et al., 2005), suggesting that they afford their host access to a vast genetic resource that can then be improved upon and made available to other organisms. Many traits that are beneficial to the host are carried by plasmids, including virulence factors, antibiotic resistance, detoxifying agents and enzymes for secondary metabolism (Philppon et al., 2002; Yates et al., 2006; Martínez, 2008; Martinez, 2009). As well as having an effect on the bacterial host, many horizontally transferred genes also code for traits that can affect the fitness of a host's neighbours. This can be either in a positive way, by producing proteins that can have a beneficial effect on the degrading enzymes of the host's neighbours (Livermore, 1995), or in a negative way, by producing substances that harm the host's neighbours, such as bacteriocins (Dykes and Hastings, 1999; Riley and Wertz, 2002; van der Ploeg, 2005; Brown et al., 2006).
In spite of their potentially positive effects on host fitness, it is important to consider that MGEs do not necessarily share the same interest as that of the host genome and can thus be considered first and foremost as infectious agents; they infect their hosts much in the same way that parasites infect their host (through contact for plasmids, or through propagules for phages), and can thus persist in spite of any costs they may impose on the host. As a result of their distinct transmission mechanisms, the interests of MGEs may not always coincide with the interests of the host genome, as they can use different means to ensure they get as many copies of themselves into the next generation (Orgel and Crick, 1980; Wagner, 2009). Whether they confer beneficial or detrimental effects on a host will depend on the selective forces exerting an effect on both the mobile element and the host chromosome.
This review will deal with the evolution of MGEs, in particular the evolutionary explanations for why they harbour the particular balance of traits they carry. Although the bulk of this review will focus on plasmids, the general results will help to understand other systems of HGT. We will focus on addressing the conditions under which certain gene classes will be favoured if transmitted horizontally. Mobile elements transmit, and are selected, at a variety of biological scales, from within the genome, to between hosts, and in larger ‘epidemiological’ patches (Figure 1). Transmission depends on the underlying ecology of the hosts, and generally increases at higher densities, whereas other factors, particularly those involved in bacterial sociality, depend strongly on the spatial structure of the population, which in turn may be influenced by the transmission rate. As such, it is essential to have a good understanding of the ecology of their bacterial hosts to better understand why certain genes are more likely to be transmitted horizontally.
An illustration of the nested nature of plasmids and their bacterial hosts. Different sub-populations may harbour different densities of plasmids. If a gene involved in cooperation is carried on a plasmid, this will affect the relatedness between individuals in a patch (because of relatedness being measured at a focal locus, in this case on a plasmid). Thus, sub-populations A and D have high plasmid relatedness, whereas sub-populations C and B have low plasmid relatedness. Relatedness is influenced by local cell density and migration between sub-populations and, in the case of loci on plasmids, relatedness changed because of the degree of horizontal gene transfer (see Box 2 and Figure 3 for details).
In this review we classify the behaviour of MGEs by their net influence on their host cell (from parasitic to mutualistic, see for example, Ferdy and Godelle, 2005) and additionally by their net influence on neighbouring cells (whether they turn their host cell into ‘helpers’ or ‘harmers’ of neighbouring cells). Hamilton (1964) proposed a way of classifying social behaviours into two types of behaviours, based on their effects on the fitness of a target individual and that of its neighbour. This classification is commonly used in social evolution theory, and, in it's contemporary form, divides social behaviours into ‘mutualism’, ‘selfishness’, ‘altruism’ and ‘spite’ (West et al., 2007b). We therefore distinguish traits that are kept within the cell (‘private’ traits, governing the parasitism–mutualism axis) from those secreted outside the cell (‘public’ traits, introducing Hamilton's social space, which govern the helping–harming axis; Hamilton, 1964; West et al., 2007b). However, for a continuum of behaviours, we divide the types of traits that can be expressed on MGEs into four types of ‘behaviours’, depending on the effects they have on the host cell and on neighbouring cells (Figure 2). We wish to go through this classification and address the types of genes that fall into these categories, and which selective pressures result in them being carried on MGEs.
Private traits: genomic parasites and mutualists
MGEs as parasites
As many prokaryotes have very large effective population sizes, any genes conferring a deleterious effect on their host will be selected against and purged from the population. However, many mobile elements do exert costs on their hosts (Diaz Ricci and Hernandez, 2000; Fox et al., 2008), raising the question as to how costly and poorly transmissible MGEs can persist within a genome (Levin and Stewart, 1977; Stewart and Levin, 1977; Bergstrom et al., 2000; Lili et al., 2007). Stewart and Levin (1977) built one of the first models to analyse plasmid persistence, and argued that plasmids could not persist under low rates of HGT. This has provoked a debate as to whether horizontal transfer is sufficient for mobile elements to persist in populations because of their costs on host fitness (see, for example, Bergstrom et al., 2000; Lili et al., 2007; Slater et al., 2008). The persistence of any parasite is fundamentally determined by its rate of horizontal transmission (Anderson and May, 1992), which can be readily measured for plasmids and has been found to vary over eight orders of magnitude among plasmids of E. coli (Dionisio et al., 2002). Although highly mobile plasmids can readily persist as molecular parasites (Bahl et al., 2007a, 2007b), the persistence of plasmids with extremely low rates of horizontal transfer cannot be readily explained in this way, given any reduction in vertical transmission imposed by the plasmid (see Box 1 ; Bergstrom et al., 2000; Dionisio et al., 2002). Rates of HGT have primarily been estimated in the laboratory, and are hard to determine for natural populations (Slater et al., 2008), yet the question of how parasitic mobile elements can persist remains an open question and important for understanding the conditions under which certain genes evolve to be transmitted horizontally. In this review we focus specifically on the evolution of horizontally transferred genes, as opposed to their persistence (readers wishing to better understand the ecological factors involved in the persistence of plasmids are directed to a recent paper on the subject by Slater et al., 2008).
Infectious elements such as plasmids are likely to face a tradeoff between horizontal transfer and vertical transmission, mediated by the costs that they impose on their hosts (Turner et al., 1998; Turner, 2004; Haft et al., 2009). Any tradeoff between HGT and costs to the host can be viewed as a form of the much-discussed virulence–transmission tradeoff (Alizon et al., 2009). However, unlike purely horizontally transmitted parasites, selection to reduce costs to the host is likely to provide additional benefits to the parasite through enhanced vertical transmission. Studies on conjugative plasmids showed that the cost of mobile elements generally decreases under selection (Bouma and Lenski, 1988; Turner et al., 1998); plasmids that exert less of a cost on their hosts have a higher representation in daughter cells. However, as an increase in vertical transfer generally comes at a cost to HGT, there will be an optimum in which gene transfer through some mix of both horizontal and vertical mechanisms is maximized (see, for example, Paulsson, 2002).
All mobile elements have mechanisms that allow them to replicate and persist within a genome. For example, class I transposons (reterotransposons) copy themselves and paste the new copy elsewhere in a genome, whereas most class II transposons (DNA transposons) cut themselves out and paste themselves elsewhere in a genome. Both plasmids and phages often come with the genetic architecture needed to replicate and transmit themselves to new hosts, either through conjugation or cell lysis. Mobile elements can be subject to resistance from the rest of the genome by several silencing or degrading mechanisms, including methylation, restriction, recognition by CRISPR, and so on (see, for example, Korona and Levin, 1993; Barrangou et al., 2007; Johnson, 2007). It is likely that hosts will evolve resistance to MGEs if they are too costly to the genome. For example, there is a large variance in the transmissibility of bacterial plasmids (Dionisio et al., 2002), and selection is likely to exert an effect upon hosts to reduce the amount of conjugation if transfer is costly. Thus, in many plasmids the expression of the conjugation machinery depends on cell density (Kozlowicz et al., 2006), or is repressed after some lag upon acquisition of the plasmid (Polzleitner et al., 1997), presumably because by then most recipients cells already contain the plasmid and the costs of conjugation offset the gains in transfer.
Gene loss is a common threat to mobile elements, such as through excision (as in the case of transposable elements) or through segregation (in the case of conjugative plasmids). Segregation occurs when no plasmids are transferred to a daughter cell during cell division. Plasmids in particular have evolved mechanisms that allow them to persist in the face of segregation. Known as plasmid ‘addiction’, these complexes come with a gene coding for a toxin, as well as a gene coding for an antitoxin (Arcus et al., 2005; Buts et al., 2005; Van Melderen and De Bast, 2009). The antitoxin is less stable than the toxin, and is broken down at a faster rate. As the addiction genes are on the plasmid, if the plasmid is lost through segregation, the toxin will remain in the cell (as it breaks down at a slower rate), but without the antitoxin the toxin kills the cell. It is worth noting that a similar principle can be observed to operate in restriction modification systems (Kobayashi, 2001). Additional and complementary hypotheses for toxin–antitoxin evolution exist (Magnuson, 2007). For example, it has also been suggested that addiction complexes have evolved not as a way for plasmids to avoid segregation, but to exclude competing plasmids (Cooper and Heinemann, 2000, 2005; Mochizuki et al., 2006). This is potentially an effective way for the plasmid to maintain itself within a population, through competition with other plasmids. However, a plasmid that codes for an addiction complex exerts a cost on the cell, which means such a plasmid will have a lower fitness with respect to other plasmids. As the plasmid is lost regardless of whether it kills the cell or not, the complex cannot be explained by invoking direct fitness benefits to the plasmid. One way that this can occur is if the dead cell is replaced by another cell carrying the addiction complex (Rankin and Brown, in preparation). As such, addiction complexes should be seen as a form of life insurance, in which in the event of the plasmid being lost through segregation, the addiction genes gain an advantage by other cells with the addiction complex replacing them with a greater probability than cells without the complex. Thus, addiction complexes can be viewed as behaving altruistically (see later section) with respect to other addiction-infected cells. This is supported by models of plasmid addiction, which find that addiction can only evolve when there is strong spatial structure (Mochizuki et al., 2006).
Consideration of the infectious nature of mobile elements highlights the selection pressures driving MGEs to optimize their own parasite function, balancing gains through transmission (vertical and horizontal) with losses through death of the host. The underlying ecology of their bacterial hosts is likely to have a large role in the evolution of mobile element virulence; if the spatial density of hosts is high, favouring HGT, mobile elements will be selected to increase their horizontal transmission, even at a cost to the host. However, if horizontal transmission is reduced to very low levels, then MGEs can only increase their fitness by coding for traits that enhance vertical transmission and for traits that are beneficial to the host (Ferdy and Godelle, 2005). Costs of mobile elements are likely to be mitigated by co-evolution with their bacterial hosts. If plasmid–chromosome co-evolution results in the evolution of reduced plasmids, which do not inflict costs on their hosts, there would be no additional fitness cost from a gene being carried on the plasmid compared with the chromosome. Small elements, such as very small plasmids, may not exert a discernible fitness effects on their host if they do not have large copy numbers. If there are no costs to a mobile element, it will be able to spread in a population through either HGT or, in the case of small populations, genetic drift. In fact, reduced costs of plasmid carriage may have been the driving force for the creation of secondary chromosomes in many bacteria. Such chromosomes often have plasmid-like features and are ubiquitous in some bacterial clades (Egan and Waldor, 2003; Slater et al., 2009). Accordingly, recent work shows that the very largest plasmids have lost mobility and acquired essential genes (Smillie et al., in preparation).
MGEs as mutualists
MGEs have been associated with the spread of a wide range of adaptive traits that enhance the fitness of their hosts. For example, plasmids commonly carry resistance genes that enable bacteria to grow in the presence of antibiotics or heavy metals, which tend to have spatially or temporarily variable distributions (Eberhard, 1990). Plasmids also have important roles in the establishment of antagonisms or mutualisms between prokaryotes and eukaryotes, such as virulence traits (Buchrieser et al., 2000), nitrogen fixation by genes coded in rhizobia symbiotic plasmids (Nuti et al., 1979) or amino acid production in Buchnera (Nuti et al., 1979; Gil et al., 2006). Phages have been shown to be important in bacterial virulence, as in the case of cholera (Faruque et al., 2005), or in mutualisms, as in the case of an aphid–bacterial association (Oliver et al., 2009).
Genes carried on plasmids may be readily transferred to the bacterial chromosome. Theoretical models suggest that constant selection pressure should favour transfer of beneficial plasmid genes to the chromosome to avoid the costs of plasmid carriage (Eberhard, 1990; Bergstrom et al., 2000). This raises the question as to why beneficial genes remain on mobile elements, rather than integrating themselves onto the chromosome. If selection pressures vary over time or space, genes that are beneficial in some environments, but not others, will be able to persist on MGEs. In a spatially structured environment, mobile elements help to facilitate the transfer of beneficial traits that have previously evolved in local populations to other sub-populations (Bergstrom et al., 2000). In addition, it has been argued that locally adapted genes that are carried on plasmids allow the persistence of these traits in the face of selective sweeps (Turner et al., 1998; Bergstrom et al., 2000), allowing for the generation of more favourable gene combinations to arise on plasmids.
Parasites and symbionts can often be observed as lying on a continuum between harming (in the case of parasites) and helping (in the case of mutualists) their partners. Mobile elements that code for beneficial genes can be seen as mutualists and should face the same evolutionary dilemmas (Ferdy and Godelle, 2005; Foster and Wenseleers, 2006; Sachs and Simms, 2007). When should a mobile element provide a benefit and when should it harm its host, and how will these evolutionary decisions in turn shape mobility? The answer to these questions are likely to depend on the details of the tradeoffs between horizontal transmission, vertical transmission and virulence (Ferdy and Godelle, 2005; Haft et al., 2009). The more an element transmits vertically, the more it will depend on the reproductive value of the host, and selection will therefore favour genes that are beneficial to the host, whereas higher HGT rates will exert an effect to the detriment of the host (subject to tradeoffs). Over long timescales, elements that harm the host are likely to be lost, and those that are beneficial to the host will likely be integrated into the host genome.
Despite the potential benefits of some plasmids to the organisms involved, sharing beneficial DNA with other members of the population can be seen as a social dilemma. The reason for the dilemma is that an organism that does not suppress the transmission of a plasmid will be benefiting its potential competitors. If transmitting a plasmid is costly, then this may be seen as an altruistic act. In this way, HGT has been suggested to be analogous to teaching in the spread of culture in humans (Riboli-Sasco et al., 2008). Behaviours learned from teaching are similar to plasmids in the sense that transfer is costly, they frequently confer a benefit on the recipient and the recipient can further transmit the information to other individuals (Riboli-Sasco et al., 2008).
MGEs as drivers of bacterial sociality
Figure 2 follows from WD Hamilton's classical categorization of social behaviours (Hamilton, 1964; West et al., 2007b, 2007c), in which social behaviours are categorized by their net lifetime direct effect on the fitness of a focal bacterium (the actor) and on neighbouring bacteria (the recipients). The resulting four behaviours are ‘selfishness’ (+/−), which confers a benefit on the actor while inflicting a cost on the social partner, ‘altruism’ (−/+), ‘spite’ (−/−) and ‘mutually beneficial behaviours’ (+/+; Hamilton, 1964; West et al., 2007c). Over the past decade, a growing body of work has been devoted to microbial sociality, and all four of these social interactions can be observed in microbial populations (Crespi, 2001; Rainey and Rainey, 2003; Griffin et al., 2004; West et al., 2006, 2007a; Xavier and Foster, 2007). We assert that when analysing social behaviours, all of these four behaviours can be observed to be encoded by horizontally transferred genes. We further argue that mobile elements, because of their effect on the local genetic structure, lend themselves to promoting cooperative social traits.
Cooperation: altruism and mutually beneficial traits
Bacteria are remarkably cooperative, producing a diversity of shared, secreted products (public goods) that can enhance growth in a diversity of challenging environments (West et al., 2006, 2007a). However, similar to any social organism, cooperative bacteria are always vulnerable to exploitation by non-producing cheats. In the absence of any kin or spatial structure, any individuals in a population that do not produce a public good will have an advantage over individuals that do, a situation referred to as the tragedy of the commons (Hardin, 1968; Rankin et al., 2007). As non-producing individuals have a fitness advantage over those that produce public goods, non-producing individuals will invade over time.
Many traits that have been shown to be involved in bacterial cooperation and virulence are coded by mobile elements, such as traits that are capable of breaking down toxins in the local environment (Knothe et al., 1983; Philppon et al., 2002; Lee et al., 2006; Ellis et al., 2007) and secreted toxins (O’Brien et al., 1984; Waldor and Mekalanos, 1996; Ahmer et al., 1999). One theoretical study (Smith, 2001) suggested that if cooperation is coded on plasmids, these plasmids will be able to reinstate cooperation by forcing non-producing strains (through infection) to start secreting the extracellular public good (Smith, 2001). However, once such a cooperative plasmid invaded a population of non-cooperating individuals, the population would still be prone to invasion from an incompatible (that is, rival) plasmid, which did not code for cooperation. Thus, over time it is likely that cooperation would break down, as the social dilemma repeats itself at the level of plasmids (Mc Ginty et al., in preparation; Nogueira et al., 2009).
Figure 1 illustrates the nested population structure of bacterial plasmids, in which plasmids are nested within bacteria, and these bacteria, in turn, are nested within patches (for example, hosts or other resource pools). As plasmids can spread horizontally within a local population, they have the potential to change the genetic structuring among individuals in local populations. Relatedness is an influential measure of population genetic structure, used in particular to decipher the direction of selection on social traits (Hamilton, 1964; Griffin et al., 2004; Rousset, 2004; West et al., 2006), with higher relatedness between individuals tending to favour the evolution of cooperative traits (Hamilton, 1964). Relatedness should properly be measured for a focal locus directly. Box 2 describes a commonly used method to calculate relatedness, and shows that relatedness at mobile loci can increase as a result of HGT. Although spatial structure, leading to increased genetic relatedness among bacterial hosts, can lead to cooperative traits on the host chromosome, they will be more strongly favoured on plasmids, as the nature of HGT can exert an effect to increase genetic relatedness even more at mobile loci (Figure 3). As such, it is expected that many genes involved in the production of cooperative traits, or public goods, will be carried by MGEs, in particular by the most transmissible conjugative plasmids.
The effect of horizontal gene transfer rate β on genetic relatedness between individuals within a patch, measured at loci with transfer rate β. The equation follows from that described in Box 2, where m=0.1, N=50 and s=0. In the absence of any HGT (that is, β=0) for the parameters chosen, r=0.079 (shown by the dotted line).
Genes coding for proteins that are secreted outside of the cell can be referred to as being part of the secretome (Ahmer et al., 1999; Sanchez et al., 2008; Nogueira et al., 2009). As proteins are costly to produce, secreting them outside of a cell has a clear cost to the focal individual. By analysing the genomes of 20 Escherichia and Shigella lineages, and also by analysing where proteins were likely to be expressed within a cell, Nogueira et al. (2009) were able to identify what genes coded for secreted proteins. The genomes were further analysed for evidence of transfer hotspots (areas in the genome more likely to be transmitted horizontally) and for identifying whether the genes coding for secreted proteins were more likely to be found on transfer hotspots or plasmids. Among genes for which localization could be predicted, it was found that 8% of chromosomal hotspot genes coded for extracellular proteins, a higher percentage than for coldspots (6%), which is the least mobile gene class. Furthermore, 15% of plasmid genes—the most mobile class of genes in the analysis—coded for extracellular proteins (Nogueira et al., 2009). This suggests that HGT is an important factor in the evolution of social behaviours in microbes (Nogueira et al., 2009).
Another example of plasmids influencing the social environment comes from the tumour-inducing plasmid in the bacterium Agrobacterium tumefaciens, a plant pathogen. For the bacterium to be virulent, it must carry a copy of the tumour-inducing plasmid, which is then inserted into the cells of plants. Once inside, the plasmid induces cell division, which creates a gall (Zupan et al., 2000). The gall releases opines that can then be used as an important source of nitrogen and energy for the bacterium (Zupan et al., 2000; White and Winans, 2007). An interesting twist in the tale is that only bacteria that are infected with the plasmid can use the opines, meaning that cooperation by plasmids only favours other individuals that contain the plasmid, making the trait a so-called ‘greenbeard’ (Gardner and West, in press).
Spiteful traits
The flipside of altruism is spite, in which an individual pays a cost to inflict another cost on another individual (Hamilton, 1970; Gardner and West, 2004a, 2004b, 2006). This is particularly common in the case of bacteria, in which individuals may produce bacteriocins, or other toxins, to kill their conspecifics (Dykes and Hastings, 1999; Riley and Wertz, 2002; Gardner et al., 2004; Ackermann et al., 2008). Spite has long been observed as a puzzling evolutionary phenomenon, as a given actor pays a net cost –c to inflict a net cost –b on another member of the population (Foster et al., 2001). Spite occurs in a number of systems (Keller and Ross, 1998; Foster et al., 2000; Gardner et al., 2004, 2007) but has most predominantly been invoked with respect to anticompetitor behaviours in bacteria (Gardner et al., 2004; West et al., 2006, 2007a; Dionisio, 2007; Brown et al., 2009a).
Spiteful extracellular products, which are costly for the producer and cause harm to other members of the population, are frequently found on mobile elements, particularly plasmids and phages (see, for example, Brown et al., 2006). In the case of bacteriocins, plasmids can carry both toxins and the corresponding genes for resistance to the toxin (Riley and Wertz, 2002). Spite can evolve if the individual exerting an effect spitefully and the individual on the receiving end of the spiteful behaviour are negatively related (that is, if r<0). In other words, as the average relatedness in a population must be r=0, the respective individuals must be less related than average, and hence damaging these individuals will increase the proportion of individuals carrying the focal actor genes. Usually this occurs under small population sizes (Gardner and West, 2004b) or if there is a way of recognizing unrelated individuals (Keller and Ross, 1998; Brown and Buckling, 2008). If bacteriocins are carried on plasmids, then this becomes easier; if a cell coding for bacteriocins lyses, it kills all members of the neighbourhood that do not bear the plasmid gene, and thus favours individuals that carry the plasmid (as they also carry resistance to the bacteriocin). As such, one may regard such genes as being ‘greenbeards’, as bacteriocinogenic individuals (carriers of the toxin-immunity gene complex) preferentially help other individuals carrying the exact same complex to survive (Gardner and West, in press). Thus, the plasmid-carried bacteriocins may be seen as analogous to the plasmid addiction complexes discussed previously, as they are mechanisms that favour the genes being on plasmids but, unlike addiction complexes (which work within the cell to kill individuals that do not carry the plasmid), they work by killing neighbouring cells.
We should expect bacteriocins to be carried by plasmids at intermediate levels of HGT or higher rates of segregation; if transfer is too high, and all individuals in a local neighbourhood carry a plasmid, then there are no non-carriers to kill. In contrast, if plasmids are too rare, then there would be insufficient toxin and subsequent killing to compensate for the fixed costs of toxin production (Chao and Levin, 1981). The targets of bacteriocins are not always the same bacterial strain (Cascales et al., 2007). This presents an additional function for bacteriocin-carrying plasmids; if they are less efficient at infecting other strains, any plasmids that produce toxins to kill those strains will have an advantage from reducing competition with the strain it can more efficiently infect. However, as for other social traits, this will very much depend on the genetic structure of the local environment.
Conclusions
Our review has highlighted the fact that a diversity of traits can be coded in mobile elements. Among this diversity, we argue that there are systematic biases towards the carriage of certain traits. It is clear that mobile elements frequently evolve to confer benefits on their bacterial hosts (see for example, Bergstrom et al., 2000; Philppon et al., 2002; Faruque et al., 2005; Slater et al., 2008; Oliver et al., 2009). It is also particularly striking that a wide range of secreted products can be found to be coded in the mobile part of the genome (Nogueira et al., 2009), highlighting the importance of mobile elements as agents of cooperation and conflict in microbial populations, and as model systems in which to study questions related to social evolution. With the growing number of bacterial and plasmid genomes becoming fully available, testing questions regarding social evolution with large genomic data sets will become increasingly feasible.
Many genes that are carried by MGEs are involved in both the evolution of antibiotic resistance and bacterial virulence. Therefore, understanding the conditions under which these genes are carried on MGEs is essential if we are to get a better understanding of how pathogenic bacteria evolve, and how potentially we can control them (Martínez, 2008; Martinez, 2009). Understanding the selective pressures that favour genes to be carried on plasmids could be useful to evaluate whether controlling plasmid transmission is an effective long-term mechanism to counter the evolution of antibiotic resistance (Amabile-Cuevas and Heinemann, 2004). The emerging importance of MGEs for the expression of bacterial cooperative phenotypes (Nogueira et al., 2009) opens up a further arena of potential biomedical application. Understanding the link between bacterial cooperation and virulence (Harrison et al., 2006) can in principle contribute to new medical intervention strategies, such as the use of genetically engineered ‘Trojan cheat’ bacteria to invade and undermine established cooperative foci of infection (Brown et al., 2009b). The realization that cooperative traits are often plasmid encoded opens the analogous possibility of introducing ‘Trojan plasmids’ to undermine the cooperative virulence of the focal population and/or drive medically useful alleles into the focal population that favours secondary mechanisms of control.
References
Achtman M (1975). Mating aggregates in Escherichia coli conjugation. J Bacteriol 123: 505–515.
Ackermann M, Stecher B, Freed NE, Songhet P, Hardt W-D, Doebeli M (2008). Self-destructive cooperation mediated by phenotypic noise. Nature 454: 987–990.
Ahmer BMM, Tran M, Heron F (1999). The virulence plasmid of Salmonella typhimurium is self-transmissible. J Bacteriol 181: 1364–1368.
Alizon S, Hurford A, Mideo N, van Baalen M (2009). Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol 22: 245–259.
Amabile-Cuevas CF, Heinemann JA (2004). Shooting the messenger of antibiotic resistance: plasmid elimination as a counter-evolutionary tactic. Drug Discov Today 9: 465–467.
Anderson RM, May RM (1979). Population biology of infectious diseases: part I. Nature 280: 361–367.
Anderson RM, May RM (1992). Infectious Diseases of Humans: Dynamics and Control. Oxford University Press: Oxford.
Arcus VL, Rainey PB, Turner SJ (2005). The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol 13: 360–365.
Bahl MI, Hansen LH, Licht TR, Sorensen SJ (2007a). Conjugative transfer facilitates stable maintenance of IncP-1 plasmid pKJK5 in Escherichia coli cells colonizing the gastrointestinal tract of the germfree rat. Appl Environ Microbiol 73: 341–343.
Bahl MI, Lars Hestbjerg H, Søren JS (2007b). Impact of conjugal transfer on the stability of IncP-1 plasmid pKJK5 in bacterial populations. FEMS Microbiol Lett 266: 250–256.
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712.
Bergstrom CT, Lipsitch M, Levin BR (2000). Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155: 1505–1519.
Bordenstein SR, Reznikoff WS (2005). Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol 3: 688–699.
Bouma JE, Lenski RE (1988). Evolution of a bacteria plasmid association. Nature 335: 351–352.
Brown SP, Buckling A (2008). A social life for discerning microbes. Cell 135: 600–604.
Brown SP, Inglis RF, Taddei F (2009a). Evolutionary ecology of microbial wars: within-host competition and (incidental) virulence. Evol Appl 2: 32–39.
Brown SP, Le Chat L, De Paepe M, Taddei F (2006). Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr Biol 16: 2048–2052.
Brown SP, West SA, Diggle SP, Griffin AS (2009b). Social evolution in micro-organisms and a Trojan horse approach to medical intervention strategies. Philos Trans R Soc London B Biol Sci 364: 3157–3168.
Buchrieser C, Glaser P, Rusniok C, Nedjari H, D’Hauteville H, Kunst F et al. (2000). The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 38: 760–771.
Bushman F (2002). Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbour University Press: Cold Spring Harbour, NY.
Buts L, Lah J, Dao-Thi M-H, Wyns L, Loris R (2005). Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci 30: 672–679.
Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K et al. (2007). Colicin biology. Microbiol Mol Biol Rev 71: 158–229.
Cascales E, Christie PJ (2003). The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1: 137–149.
Chao L, Levin BR (1981). Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA 78: 6324–6328.
Chen I, Dubnau D (2004). DNA uptake during bacterial transformation. Nat Rev Microbiol 2: 241–249.
Cooper TF, Heinemann JA (2000). Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competiting plasmids. Proc Natl Acad Sci USA 97: 12643–12648.
Cooper TF, Heinemann JA (2005). Selection for plasmid post-segregational killing depends on multiple infection: evidence for the selection of more virulent parasites through parasite-level competition. Proc R Soc London B Biol Sci 272: 403–410.
Crespi BJ (2001). The evolution of social behavior in microorganisms. Trends Ecol Evol 16: 178–183.
de la Cruz F, Frost LS, Meyer RJ, Zechner E (2009). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34: 18–40.
Delsuc F, Brinkmann H, Philippe H (2005). Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6: 361–375.
Diaz Ricci JC, Hernandez ME (2000). Plasmid effects on Escherichia coli metabolism. Crit Rev Biotechnol 20: 79–108.
Dionisio F (2007). Selfish and spiteful behaviour through parasites and pathogens. Evol Ecol Res 9: 1199–1210.
Dionisio F, Matic I, Radman M, Rodrigues OR, Taddei F (2002). Plasmids spread very fast in heterogeneous bacterial communities. Genetics 162: 1525–1532.
Dykes GA, Hastings JW (1999). Selection and fitness in bacteriocin-producing bacteria. Proc R Soc London B Biol Sci 264: 683–687.
Eberhard WG (1990). Evolution in bacterial plasmids and levels of selection. Q Rev Biol 65: 3–22.
Egan ES, Waldor MK (2003). Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114: 521–530.
Ellis RJ, Lilley AK, Lacey SJ, Murrell D, Godfray HCJ (2007). Frequency-dependent advantages of plasmid carriage by Pseudomonas in homogeneous and spatially structured environments. ISME J 1: 92–95.
Faruque SM, Naser IB, Islam MJ, Faruque ASG, Ghosh AN, Nair GB et al. (2005). Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc Natl Acad Sci USA 102: 1702–1707.
Ferdy JB, Godelle B (2005). Diversification of transmission modes and the evolution of mututalism. Am Nat 166: 613–627.
Foster KR, Ratnieks FLW, Wenseleers T (2000). Spite in social insects. Trends Ecol Evol 15: 469–470.
Foster KR, Wenseleers T (2006). A general model for the evolution of mutualisms. J Evol Biol 19: 1283–1293.
Foster KR, Wenseleers T, Ratnieks FLW (2001). Spite: Hamilton's unproven theory. Ann Zool Fennici 38: 229–238.
Fox RE, Zhong X, Krone SM, Top EM (2008). Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J 2: 1024–1039.
Frost LS, Leplae R, Summers AO, Toussaint A (2005). Mobile genetic elements: the agents of open source evolution. Nat Rev Micro 3: 722–732.
Gardner A, Hardy ICW, Taylor PD, West SA (2007). Spiteful soldiers and sex ratio conflict in polyembryonic parasitoid wasps. Am Nat 169: 519–533.
Gardner A, West SA (2004a). Spite among siblings. Science 305: 1413–1414.
Gardner A, West SA (2004b). Spite and the scale of competition. J Evol Biol 17: 1195–1203.
Gardner A, West SA (2006). Spite. Curr Biol 16: R662–R664.
Gardner A, West SA (2010). Greenbeards. Evolution 64: 25–38.
Gardner A, West SA, Buckling A (2004). Bacteriocins, spite and virulence. Proc R Soc London B 271: 1529–1535.
Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ et al. (2005). Re-evaluating prokaryotic species. Nat Rev Micro 3: 733–739.
Gil G, Sabater-Muñoz B, Perez-Brocal V, Silva FJ, Latorre A (2006). Plasmids in the aphid endosymbiont Buchnera aphidicola with the smallest genomes. A puzzling evolutionary story. Gene 370: 17–25.
Goldenfeld N, Woese C (2007). Biology's next revolution. Nature 445: 369.
Griffin AS, West SA, Buckling A (2004). Cooperation and competition in pathogenic bacteria. Nature 430: 1024–1027.
Hacker J, Carniel E (2001). Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep 2: 376–381.
Haft RJF, Mittler JE, Traxler B (2009). Competition favours reduced cost of plasmids to host bacteria. ISME J 3: 761–769.
Hamilton WD (1964). The genetical evolution of social behaviour. I & II. J Theor Biol 7: 1–52.
Hamilton WD (1970). Selfish and spiteful behaviour in an evolutionary model. Nature 228: 1218–1220.
Hardin G (1968). The tragedy of the commons. Science 162: 1243–1248.
Harrison F, Browning LE, Vos M, Buckling A (2006). Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol 4: 21.
Johnson LJ (2007). The genome strikes back: the evolutionary importance of defence against mobile elements. Evol Biol 34: 121–129.
Keller L, Ross KG (1998). Selfish genes: a green beard in the red fire ant. Nature 394: 573–575.
Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S (1983). Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11: 315–317.
Kobayashi I (2001). Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res 29: 3742–3756.
Korona R, Levin BR (1993). Phage-mediated selection for restriction-modification. Evolution 47: 565–575.
Kozlowicz BK, Shi K, Gu ZY, Ohlendorf DH, Earhart CA, Dunny GM (2006). Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol Microbiol 62: 958–969.
Kurland CG, Canback B, Berg OG (2003). Horizontal gene transfer: a critical view. Proc Natl Acad Sci USA 100: 9658–9662.
Lederberg J, Tatum EL (1946). Gene recombination in E. coli. Nature 158: 558.
Lee K, Lee M, Shin JH, Lee MH, Kang SH, Park AJ et al. (2006). Prevalence of plasmid-mediated AmpC β-Lactamases in Escherichia coli and Klebsiella pneumoniae in Korea. Microb Drug Resist 12: 44–49.
Lehmann L, Feldman MW, Foster KR (2008). Cultural transmission can inhibit the evolution of altruistic helping. Am Nat 172: 12–24.
Lehmann L, Keller L (2006). The evolution of cooperation and altruism—a general framework and a classification of models. J Evol Biol 19: 1365–1376.
Lerat E, Ochman H (2004). Psi-Phi: exploring the outer limits of bacterial pseudogenes. Genome Res 14: 2273–2278.
Levin BR, Stewart FM (1977). Probability of establishing chimeric plasmids in natural populations of bacteria. Science 196: 218–220.
Lili LN, Britton NF, Feil EJ (2007). The persistence of parasitic plasmids. Genetics 177: 399–405.
Livermore DM (1995). Beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev 8: 557–584.
Magnuson RD (2007). Hypothetical functions of toxin-antitoxin systems. J Bacteriol 189: 6089–6092.
Martínez JL (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321: 365–367.
Martinez JL (2009). The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc R Soc London B 276: 2521–2530.
May RM, Anderson RM (1979). Population biology of infectious diseases: part II. Nature 280: 455–461.
Mochizuki A, Yahara K, Kobayashi I, Iwasa Y (2006). Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics 172: 1309–1323.
Nogueira T, Rankin DJ, Touchon M, Taddei F, Brown SP, Rocha EP (2009). Gene mobility drives the evolution of bacterial cooperation and virulence. Curr Biol 19: 1683–1691.
Norman A, Hansen LH, Sorensen SJ (2009). Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc London B 364: 2275–2289.
Nuti MP, Lepidi AA, Prakash RK, Schilperoort RA, Cannon FC (1979). Evidence for nitrogen fixation (nif) genes on indigenous Rhizobium plasmids. Nature 282: 533–535.
O’Brien AD, Newland JW, Miller SF, Holmes RK, Smith HW, Formal SB (1984). Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226: 694–696.
Ochman H, Lawrence JG, Groisman EA (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304.
Oliver KM, Degnan PH, Hunter MS, Moran NA (2009). Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325: 992–994.
Orgel LE, Crick FHC (1980). Selfish DNA: the ultimate parasite. Nature 284: 604–607.
Paulsson J (2002). Multileveled selection on plasmid replication. Genetics 161: 1373–1384.
Philppon A, Arlet G, Jacoby GA (2002). Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother 46: 1–11.
Polzleitner E, Zechner EL, Renner W, Fratte R, Jauk B, Högenauer G et al. (1997). TraM of plasmid R1 controls transfer gene expression as an integrated control element in a complex regulatory network. Mol Microbiol 25: 495–507.
Rainey PB, Rainey K (2003). Evolution of cooperation and conflict in experimental bacterial populations. Nature 425: 72–74.
Rankin DJ, Bargum K, Kokko H (2007). The tragedy of the commons in evolutionary biology. Trends Ecol Evol 22: 643–651.
Rasched I, Oberer E (1986). Ff coliphages: structural and functional relationships. Microbiol Rev 50: 401–427.
Riboli-Sasco L, Brown SP, Taddei F (2008). Why teach? The evolutionary origins and ecological consequences of costly information transfer. In: Ettorre P, Hughes D (eds). Communication Among Social Organisms. Oxford University Press.
Riley MA, Wertz JE (2002). Bacteriocins: evolution, ecology and application. Annu Rev Microbiol 56: 117–137.
Rousset F (2004). Genetic Structure and Selection in Subdivided Populations. Princeton University Press: Princeton.
Sachs JL, Mueller UG, Wilcox TP, Bull JJ (2004). The evolution of cooperation. Q Rev Biol 79: 133–160.
Sachs JL, Simms EL (2007). Pathways to mutualism breakdown. Trends Ecol Evol 21: 585–592.
Sanchez B, Bressollier P, Urdaci MC (2008). Exported proteins in probiotic bacteria: adhesion to intestinal surfaces, host immunomodulation and molecular cross-talking with the host. FEMS Immunol Med Microbiol 54: 1–17.
Slater FR, Bailey MJ, Tett AJ, Turner SL (2008). Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol Ecol 66: 3–13.
Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW et al. (2009). Genome sequences of three agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol 191: 2501–2511.
Smith J (2001). The social evolution of bacterial pathogenesis. Proc R Soc London B Biol Sci 268: 61–69.
Sorensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Micro 3: 700–710.
Stewart FM, Levin BR (1977). The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics 87: 209–228.
Thomas CM, Nielsen KM (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Micro 3: 711–721.
Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P et al. (2009). Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344.
Turner PE (2004). Phenotypic plasticity in bacterial plasmids. Genetics 167: 9–20.
Turner PE, Cooper VS, Lenski RE (1998). Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52: 315–329.
van der Ploeg JR (2005). Regulation of Bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J Bacteriol 187: 3980–3989.
Van Melderen L, De Bast MS (2009). Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet 5: e1000437.
Wagner A (2009). Transposable elements as genomic diseases. Mol Biosyst 5: 32–35.
Waldor MK, Mekalanos J (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914.
West SA, Diggle SP, Buckling A, Gardner A, Griffin AS (2007a). The social lives of microbes. Ann Rev Ecol, Evol Syst 38: 53–77.
West SA, Griffin AS, Gardner A (2007b). Evolutionary explanations for cooperation. Curr Biol 17: R661–R672.
West SA, Griffin AS, Gardner A (2007c). Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. J Evol Biol 20: 415–432.
West SA, Griffin AS, Gardner A, Diggle SP (2006). Social evolution theory for microorganisms. Nat Rev Microbiol 4: 597–607.
White CE, Winans SC (2007). Cell-cell communication in the plant pathogen Agrobacterium tumefaciens. Philos Trans R Soc London B Biol Sci 362: 1135–1148.
Xavier JB, Foster KR (2007). Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA 104: 876–881.
Yates CM, Shaw DJ, Roe AJ, Woolhouse MEJ, Amyes SGB (2006). Enhancement of bacterial competitive fitness by apramycin resistance plasmids from non-pathogenic Escherichia coli. Biol Lett 2: 463–465.
Zinder ND, Lederberg J (1952). Genetic exchange in Salmonella. J Bacteriol 64: 679–699.
Zupan J, Muth TR, Draper O, Zambryski P (2000). The transfer of DNA from agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J 23: 11–28.
Acknowledgements
We are grateful to the Swiss National Science Foundation (Grants 31003A-125457 and PZ00P3-121800 to DJR) and the University of Zürich Forschungskredit (DJR), the CNRS and the Institut Pasteur (EPCR) and the Wellcome Trust (Grant 082273/Z/07/Z to SPB) for funding. We also thank Sorcha Mc Ginty and Ben Raymond for comments on the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Rankin, D., Rocha, E. & Brown, S. What traits are carried on mobile genetic elements, and why?. Heredity 106, 1–10 (2011). https://doi.org/10.1038/hdy.2010.24
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.24
Keywords
This article is cited by
-
Engineering intelligent chassis cells via recombinase-based MEMORY circuits
Nature Communications (2024)
-
Horizontal gene transfer is predicted to overcome the diversity limit of competing microbial species
Nature Communications (2024)
-
Whole genome sequence and comparative genomics analysis of multidrug-resistant Staphylococcus xylosus NM36 isolated from a cow with mastitis in Basrah city
Journal of Genetic Engineering and Biotechnology (2023)
-
Genome-associations of extended-spectrum ß-lactamase producing (ESBL) or AmpC producing E. coli in small and medium pig farms from Khon Kaen province, Thailand
BMC Microbiology (2022)
-
Comparative analysis of Streptococcus suis genomes identifies novel candidate virulence-associated genes in North American isolates
Veterinary Research (2022)