Abstract
Copy neutral segments with allelic homozygosity, also known as regions of homozygosity (ROHs), are frequently identified in cases interrogated by oligonucleotide single-nucleotide polymorphism (oligo-SNP) microarrays. Presence of ROHs may be because of parental relatedness, chromosomal recombination or rearrangements and provides important clues regarding ancestral homozygosity, consanguinity or uniparental disomy. In this study of 14 574 consecutive cases, 832 (6%) were found to harbor one or more ROHs over 10 Mb, of which 651 cases (78%) had multiple ROHs, likely because of identity by descent (IBD), and 181 cases (22%) with ROHs involving a single chromosome. Parental relatedness was predicted to be first degree or closer in 5%, second in 9% and third in 19%. Of the 181 cases, 19 had ROHs for a whole chromosome revealing uniparental isodisomy (isoUPD). In all, 25 cases had significant ROHs involving a single chromosome; 5 cases were molecularly confirmed to have a mixed iso- and heteroUPD15 and 1 case each with segmental UPD9pat and segmental UPD22mat; 17 cases were suspected to have a mixed iso- and heteroUPD including 2 cases with small supernumerary marker and 2 cases with mosaic trisomy. For chromosome 15, 12 (92%) of 13 molecularly studied cases had either Prader–Willi or Angelman syndrome. Autosomal recessive disorders were confirmed in seven of nine cases from eight families because of the finding of suspected gene within a ROH. This study demonstrates that ROHs are much more frequent than previously recognized and often reflect parental relatedness, ascertain autosomal recessive diseases or unravel UPD in many cases.
Similar content being viewed by others
Introduction
Oligonucleotide single-nucleotide polymorphism array (oligo-SNP) is an important diagnostic tool to identify both copy number variants (CNVs) and regions of homozygosity (ROHs) in patients with a wide variety of clinical indications. These ROHs are often unexpected and in many cases the clinical significance remains to be determined. Long contiguous ROHs may arise by a number of mechanisms. These include parental relatedness (or consanguinity) and chromosomal recombination or rearrangements. Depending on chromosomal distribution and cumulative extent, it may indicate background ancestral homozygosity, uniparental disomy (UPD) or parental consanguinity.1, 2, 3, 4
Small random ROHs exist in all populations, including outbred cosmopolitan ones, and are believed to be a reflection of recombination rates and population history. A recent study using HapMap trios aimed at determining the prevalence of UPD in the general population suggested the existence of segmental UPD in 1 per 173 births (∼0.6%) in the random population.5 Such regions are usually <4 Mb as a contiguous stretch of the genome, particularly in outbred cosmopolitan populations according to a study of European populations.6 In rare instances, ROH over 10 Mb as a single contiguous stretch has also been encountered and reported in certain individuals from different populations (Japan and Nigeria).3 In addition to their clinical significance, the presence and extent of ROHs in a community or population mirrors the genetic structure and diversity of the population. It is now recognized that the effect of ROHs is not only significant for monogenic recessive disorders, but there is a role for recessive variants in complex genetic disorders.7, 8, 9 When the regions with allelic homozygosity occur on multiple chromosomes, autozygosity resulting from inheritance of the genomic segment from parents who inherited it from a common ancestor is the most likely cause (a situation referred to as identity by descent (IBD)). In contrast, when such regions are restricted to a single chromosome, the possibility of UPD should be considered.
A recent clinical report of ∼3000 cases studied by SNP array identified 59 (2%) cases with ROHs1 that predicted the degree of parental relatedness and aided the diagnosis of autosomal recessive diseases in two families; however, cases with UPD were not documented in this study. Another study of over 13 000 samples with developmental delay focused on the study of UPD only, with identification of 16 cases with isoUPD, 19 cases with hetero–isoUPD and 3 cases with segmental UPD.4 There was no clinical study that provided an overall review of the finding of ROHs for all these three categories, that is, parental relatedness, autosomal recessive disorders and UPD, and this may be because of a huge variation for the threshold for reporting ROHs between laboratories,10 and a lack of recommendations regarding the standards and guidelines for documenting suspected consanguinity.11
In this study, the largest and most comprehensive series reported so far, we report the finding of 832 (6%) cases with ROH from 805 families from 14 574 clinical cases referred to our laboratory: 651 (78%) cases with multiple ROHs and likely due to parental relatedness, and 181 (22%) cases with single-chromosome involvement. The data presented here represent the largest single collection of cases analyzed by oligo-SNP arrays leading to identification of a significant number of cases harboring clinically significant ROHs. This study further demonstrates that the identification of ROHs, in addition to CNVs, is much more frequent than previously recognized. Moreover, identification of ROHs is beginning to unravel clinically significant uniparental disomy and homozygosity for recessive disease gene mutations that would otherwise not have been suspected or identified.
Materials and methods
Patients
Patients with a broad range of clinical indications including intellectual disability, developmental delay, multiple congenital anomalies, dysmorphic features and pervasive developmental disorders were referred to our laboratory for oligo-SNP array studies. Over 85% of cases were referred for one or more of the above clinical indications. A small percentage of cases were studied by oligo-SNP arrays following normal conventional cytogenetic studies (karyotype and/or locus-specific FISH). The data were compiled from consecutive specimens that were referred to Nichols Institute at San Juan Capistrano over 2 years. The patients were majorly from general population in the United States, with <5% from Mexico and other countries.
Microarray analysis and homozygosity (HMZ) screening threshold setting (oligo-SNP)
Genomic DNA extracted from whole blood was utilized for analysis on either the Genome-Wide Human SNP Array 6.0 (40% of cases) or the CytoScan HD array (60% of cases) (Affymetrix, Santa Clara, CA, USA). The SNP array 6.0 contains 1.8 million genetic markers, including ∼906 600 SNPs and 946 000 probes for the detection of copy number variation. The CytoScan HD array has more than 2.67 million probes, including 1.9 million nonpolymorphic copy number probes and 750 000 SNP probes. The density of SNP coverage between the two array platforms is similar; the mean SNP probe spacing is 3 kb and 3.6 kb for SNP 6.0 and Cytoscan HD, respectively. Genomic coordinates are based upon genome build 37/hg19 (2009). Hybridization, data extraction and analysis were performed as per the manufacturer’s protocols. The Affymetrix Chromosome Analysis Suite (ChAS) Software version 1.0 or 2.0 was used for data analysis, review and reporting. The screening threshold used for length of ROH detection was set at 5 Mb; additional stringent parameters used to detect only true ROH included a minimum of 50 contiguous homozygous SNPs within a window, and <5 contiguous heterozygous SNPs (within a 100-kb interval). Regions with sparse SNP density were carefully evaluated to exclude false ROH calls. Copy number data for all ROH segments were carefully evaluated to rule out copy number losses (to rule out ROH due to hemizygosity). Although no consensus opinion exists regarding ROH size cutoffs, the choice of 5 Mb as the cutoff value is primarily derived from the study of European populations and recent data from other laboratories.6, 10 To avoid underestimation of the amount of ROH, the screening threshold was often varied so as not to miss ROHs just below the 5 Mb cutoff. ROH was reported as the presence of one segment of homozygosity >10 Mb or at least two ROHs >5 Mb in each region (with two or more segments on two or more chromosomes classified as IBD; and all segment(s) within one chromosome classified as possible UPD).
Quantifying ROHs and classification of degree of consanguinity
The amount of coverage of human genome in the oligo-SNP array was estimated by using the most telomeric probe for each chromosome on Cytoscan software and subtracting 180 Mb for the regions with heterochromatin around the centromeres; this was estimated to be 2700 Mb for autosomes. Except for the recurrent ROH segments, all other ROHs >5 Mb were included in the reporting. The classification of degree of consanguinity was based on the 95% confidence interval (CI) obtained using a methodology similar to the previous study from Sund et al.1 CIs were calculated for five inbreeding percentages (25, 12.5, 6.25, 3.125 and 1.5625%) and for a total of 533 autosomal segments of 5 Mb each with SNP coverage.1 Percentage homozygosity values (95% CI) obtained were very similar to that reported by Sund et al1 (over 28.7% is indicative of parents being first-degree relatives or closer; 21.3–28.7% first; 15.3–21.3% first or second; 9.7–15.3% second; 8.3–9.7% second or third; 4.4–8.3% third; 4.2–4.4% third or fourth; 2.4–4.2% fourth; 2.0–2.4% fourth or fifth; 0.5–2.0% fifth).
Searching for potential candidate genes in ROHs for autosomal recessive disorders
Gene content and phenotype associations for ROH segments were evaluated by utilizing the web-based Genomic Oligoarray and SNP array evaluation tool, developed and made available by Wierenga et al.12 By entering the genomic coordinates of all ROHs in this tool (www.ccs.miami.edu/ROH), a list of the OMIM genes mapping to those particular regions, and their associated (recessive) disorders, was generated for review.12
Results
Recurrent ROHs
Five ROHs were recurrently identified in our patient population (and normal parents in many cases) and considered as polymorphic variants and not included in the reports and calculation: 3p21.31-p21.1 (chr3.hg19:g.47 586 513_53 002 150), 11p11.2-p11.12 (chr11.hg19:g.46 304 337_51 563 636), 16p11.2-p11.1 (chr16.hg19:g.29 194 414_35 220 544), Xp11.23-p11.22 (chrX.hg19:g.48 354 880_54 540 061) and Xq11.1q12 (chrX.hg19:g.61 932 503_67 123 772).
ROH due to IBD is a frequent finding in oligo-SNP array analysis
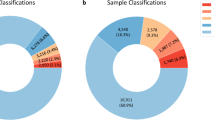
Of the 14 574 cases analyzed by oligo-SNP arrays, 9759 cases (67%) were neutral for copy number, 1453 cases (10%) had clinically significant CNVs and 3362 cases (23%) had copy number variants of unclear significance (VOUS) (Figure 1a). A total of 832 cases (6%) from 805 families had one or more reportable ROHs (Figure 1b): 548 cases with normal copy number, 79 cases with clinically significant CNVs and 205 cases with VOUS. These cases included sibling studies of a total of 50 patients from 23 families.
(a) A total of 14 574 cases were analyzed by oligo-SNP arrays: 9759 cases (67%) had a neutral copy number, 3362 cases (23%) had variants of unclear significance (VOUS) and 1453 cases (10%) had clinically significant copy number variations (CNVs). (b) In all, 832 cases (6%) had one or more reportable ROHs: 651 (78%) cases were interpreted as arising because of identity by descent (IBD), and 181 (22%) cases were suspected or confirmed as resulting from UPD. In cases due to IBD, 68% are copy number neutral, 25% had VOUS and 7% had clinically significant CNVs. In the cases with suspected or confirmed UPD, 58% are copy number neutral, 24% had VOUS and 18% had clinically significant CNVs.
Of the 832 cases with ROHs, 651 (78%) cases had multiple ROHs from more than one chromosome and were interpreted as indicative of IBD. The cumulative ROHs ranged from 10.2 to 1096 Mb in size (mean=135 Mb; median=67 Mb; Figure 2). An estimate of the parental relatedness was determined: 31 (5%) cases with a first degree or closer kinship between the parents (see, Supplementary Figure 1); 58 (9%) cases second degree or closer; 130 (20%) cases third degree or closer (see, Supplementary Figure 2); 114 (17%) cases fourth degree or closer; 318 (49%) cases fifth degree or closer (Table 1a).
Total number of cases in each of the five inbreeding groups. The X axis represents the homozygosity percentage values and the corresponding cumulative length for the five groups (95% confidence interval). The Y axis represents the total number of cases in each group (Table 1a).
Sibling studies for 50 cases from 23 families (Table 1b) showed relatively concordant results in prediction of parental relatedness in 13 families, such as family 3 (three siblings each with ∼876 Mb ROH or 32% HMZ in total) and family 20 with 289 Mb (10.7% HMZ) and 329 Mb ROHs (12.2% HMZ in two siblings). However, in some families, siblings may both have significant ROHs, but it did not predict the same degree of parental relatedness; for instance, in families 1 and 2, the percentage HMZ was 7.4–11.1% and 4.1–13.5%, respectively. Such discordant IBD predictions were detected in the siblings from families 1, 2, 8, 9, 10, 11, 12, 14, 16, 18 and 19.
Almost one-quarter of cases with ROHs arising from one single chromosome are because of suspected or confirmed isoUPD, mixed iso- and heteroUPD or segmental UPD
In 181 of 832 cases (22%), ROH from a single chromosome was detected. In 137 cases, the ROH was <25% of the size of the chromosome involved, raising the possibility of segmental UPD. Of the remaining 44 cases, 19 had ROH for a whole chromosome (uniparental isodisomy or isoUPD): 7 involving chromosome 15, 3 from chromosome 2, 2 from chromosome 7 and 1 each from chromosomes 1, 5, 8, 9, 16, 19 and 20 (Table 2a) (see, eg, Supplementary Figure 3). One case with UPD5 was associated with a small, mosaic supernumerary marker chromosome 5 (1.3 Mb in size) (18 of 20 cells), and one case with UPD8 and a familial nonmosaic marker chromosome 8 (6.1 Mb). In the remaining 25 cases, ROHs did not involve the whole chromosome (Table 2b) and included 8 from chromosome 15 (see, eg, Supplementary Figure 4) and 4 from chromosome 9. Concurrent ROHs and mosaic trisomy for chromosomes 9 and 18 were seen in 2 cases, respectively, and could most likely be because of trisomy rescue.13 Concurrent ROHs (mixed iso- and heteroUPD) and small supernumerary marker chromosomes were seen in two cases, from chromosomes 1 and 4, respectively.14
The majority of cases with ROHs from chromosome 15 are due to UPD15
Among the 15 cases with ROHs from chromosome 15 alone (Table 2c), 7 had isoUPD for the entire chromosome 15 and 8 had ROH from 13 to 47 Mb (Table 2c, Supplementary Figure 4). All 7 cases with ROH of whole chromosome 15, and 5 of 8 cases (63%) cases with segmental ROH 15 were molecularly confirmed to be clinically PWS or AS. Please note that both cases with ROH of only 13 and 16 Mb (16 and 20% of the size of whole chromosome 15) had Prader–Willi syndrome (Table 2b, cases 9 and 10; Table 2c, cases 10 and 11).
UPD11 in cases with ROHs from chromosome 11 alone
Among the 13 cases with ROHs from chromosome 11 alone, none involve the whole chromosome. One case with a clinical indication of hemihypertrophy and hypoglycemia in the neonatal period was revealed to have a segmental ROH in a mosaic state that was ∼15 Mb in size (11p15.2-pter; arr[hg19] 11p15.5p15.2(198 510–15 029 576) × 2 mos hmz; Supplementary Figure 5). This segment includes the imprinted domain at 11p15.5 associated with Beckwith–Wiedemann syndrome (BWS, OMIM #130650). Methylation-sensitive multiplex ligation-dependent probe amplification (MLPA) performed elsewhere showed hypermethylation of IC1 (H19) and hypomethylation of IC2 (LIT1), consistent with mosaic paternal UPD11 and a clinical diagnosis of BWS.
Autosomal recessive disorders unmasked by ROHs
In nine patients (from eight families), identification of significant ROH followed by analysis of ROH segments for disease-causing genes helped molecular diagnosis in seven (Table 3). (Cases 1 and 2). The 3-year-old twin boy and girl with short stature and hydrocephalus, both with little response to growth hormone, were diagnosed to have Laron dwarfism. A homozygous c.181C>T transition, predicted to lead to a premature stop codon in the growth hormone receptor gene GHR, NM_000163.4:c.181C>T [p.(Arg61*)], was confirmed (OMIM #262500; growth hormone receptor, GHR at 5p13.1-p12). (3) A 3-year-old boy with primary microcephaly (FOC 43.5 cm at 3 years of age), developmental delay, speech delay, close-spaced eyes, epicanthal folds, downslanting palpebral fissures, large ears and smooth philtrum. A homozygous deletion of ∼250 kb was found at 8p23.2-p23.1 and within a ROH, chr8.hg19:g.(6060654_6061169)_(6310738_6317266)del. The deleted region includes the microcephalin gene, MCPH1, and most likely resulting in primary autosomal recessive microcephaly 1 (OMIM #251200, MCPH1 at 8p23.2) (Hemmat et al., manuscript in preparation, 2014). (4) An 18-day-old male baby with organic aciduria and elevated C5OH was diagnosed with 3-methylglutaconic aciduria, type I (OMIM #250950; AU RNA binding protein/enoyl-CoA hydratase (AUH), at 9q22.31). A homozygous missense transition mutation (NM_001698.2:c.373C>T, [p.(Arg125Trp)]) was identified in the AU-specific RNA binding protein (AUH) gene, encoding a mitochondrial protein.12 This sequence change affects a highly conserved amino acid residue located in a functional domain of the AUH protein. The homozygous c.373C>T change appears to be deleterious using several in silico prediction tools (SIFT, Polyphen-2, MutationTaster) and in the context of this patient’s clinical presentation it is interpreted as most likely pathogenic. (5) A 23-day-old baby male with coarse features, unusual palate, pectus excavatum, long fingers, contractures and camptodactyly was suspected to have mucolipidosis type II (OMIM #252500) and strongly suspected to harbor homozygous mutation in the gene N-acetylglucosamine-1-phosphate transferase, alpha/beta subunits (GNPTAB, at 12q23.2). (6) A 6-month-old male child with partial seizures, mild white matter hypoplasia with corpus callosum and enlargement of the lateral ventricles shown in brain MRI, abnormal EEG, hypotonia, wide inner canthal distance, widely spaced nipples and wide nasal bridge was suspected to have bare lymphocyte syndrome type II (OMIM #209920, regulatory factor X-associated protein (RFXAP) at 13q13.3). A homozygous nonsense mutation in exon 1 of the RFXAP gene predicted to result in a premature stop codon was identified (NM_000538.3:c.323T>A, [p.(Leu108*)]). (7) A 7-year-old female suspected to have Bardet–Biedl syndrome 4 (OMIM #209900, BBS4 gene at 15q23). (8 and 9). An additional two cases with a confirmed recessive disorder with one having segmental UPD9 and the second with whole-chromosome UPD16, respectively (Table 3, cases 8 and 9). Case 8 is a 5-year-old girl with elevated citrullin, spastic quadriparesis, developmental delay and found to have autosomal recessive citrullinemia (OMIM #215700, argininosuccinate synthetase 1 (ASS1) gene at 9q34.11) harbored a complex abnormality involving 9q including homozygosity for the 9q21.33-qter segment. A homozygous missense mutation in the ASS1 gene (NM_000050.4:c.571G>A, [p.(Glu191Lys)]) was identified and found to have been inherited from a heterozygous carrier parent (case 4, Table 2b). This mutation has been previously described in at least two patients with autosomal recessive citrullinemia.15, 16 Case 9 is a 24-day-old female baby with whole-chromosome UPD16 suspected to have Bardet–-Biedl syndrome 2 (OMIM #209900 and 606151, BBS2 gene at 16q12.2). A homozygous frameshift mutation in exon 14 of BBS2 gene was identified and predicted to lead to a premature stop codon (NM_031885.3:c.1770delT, [p.(Phe590Leufs*8]). One parent of the proband was identified to be a heterozygous carrier for the identical mutation.
Discussion
Implementation of oligo-SNP arrays has enabled simultaneous interrogation of genomic DNA for copy number variations and copy neutral regions of allelic homozygosity. The availability of this information proves invaluable in providing diagnostic information critical to the clinical evaluation of patients with constitutional genetic disorders. The cumulative extent of ROHs in any given case is determined by multiple factors such as parental relatedness, ethnicity, recombination events and chromosomal aberrations. The significance of such findings and deriving clues towards molecular testing and genotype–phenotype correlations necessitates critical evaluation of the array data in each individual case.
Currently, there is no consensus regarding the threshold for identification of runs or ROHs in constitutional cases.10 The choice of 5 Mb as the cutoff value for identification of ROHs is primarily derived from study of European populations, demonstrating that outbred individuals rarely carried ROHs over 4 Mb in size.6 However, the cutoff value for other populations is likely to be different.17 In this study, 832 (6% of 14 574) cases were identified to harbor one or more ROHs, and this is much more frequent than previously reported.1 It may be because of a lower but appropriate size threshold, a larger case series or sampling of populations in this study.
The classification of degree of consanguinity for 651 cases was based on the 95% CI calculated following a methodology similar to that used in a previous small study of 59 patients.1 The significant agreement in the predicted percentage homozygosity for the five degrees of inbreeding (consanguinity) provides strong credibility to the methodology used and, additionally, lays the basis for reaching a consensus guideline for evaluation, interpretation and reporting of ROH. It is to be noted that, despite agreement in homozygosity percentage intervals for calculating parental relatedness, the frequency of cases for each of the five degrees was somewhat different. This can be attributed to a very large number of cases in this study that would reduce the possibility of a chance occurrence. Our data set includes close to 11 times more cases with IBD (651/59), thus strengthening our predictions. There is remote likelihood that the calculated frequencies may not reflect the true degree of consanguinity for each particular family as it has been demonstrated that the frequency of consanguinity is diverse among populations, thus leading to a varied size of extended tracts of homozygosity.3, 17
In addition to providing an estimate for parental relatedness in cases with IBD, one surprising revelation was a significant number of cases (N=31; 5%) wherein parental relatedness could be assessed to be of the first degree or closer. This level of cumulative homozygosity of 25% of the genome or greater adds a new dimension to the clinical evaluation and counseling process. It is increasingly becoming a challenge about how to respond to and counsel for such results, and a need for genetics professionals to have a guideline has been requested.18 The ethical, legal and social outcomes that these cases present and the approaches to dealing with the same are still evolving.18, 19
In the sibling studies, the result showed only 13 of 23 families (∼57%) had a concordant result in prediction of parental relatedness. Although in some families such as family 3 (Table 1b), three siblings had almost exact amount of ROHs (∼876 Mb), for other families, ROH from one sibling alone will not reliably predict the degree of relatedness between parents. For instance, in family 10, both siblings had a very similar clinical phenotype, but the ROHs for each sibling was discrete (31 vs 395 Mb), thus precluding easy genotype–phenotype correlation, or raising the possibility of nonpaternity.
Another aspect of this study is the frequency of identification of UPD (in more than 40 cases) including isoUPD (19 cases), segmental UPD (2 cases) and iso- and heteroUPD (22 cases). The major drawback to having the inclusion criteria set for at least two ROHs on two separate chromosomes, as suggested by Sund et al,1 is likely to increase the risk of missing single-chromosomal ROH that may be due to combined iso- and heteroUPD. We demonstrated that ROHs only from a single chromosome is frequently encountered and can involve any chromosome (Table 2a). Large chromosomes such as chromosomes 1–3 and chromosomes with imprinted genes are more frequently identified than other chromosomes.
The ratio of both pure iso- and iso/heteroUPD15 in cases with ROH restricted to chromosome 15 is much higher than the other chromosomes (Tables 2b and c). In the cases with ROHs from chromosome 15, follow-up molecular studies (in 13 of 15 cases) confirmed UPD15 in 12 of 13 cases and helped reach a clinical diagnosis of PWS or AS. Two cases with small ROHs (<25% of the whole chromosome) were molecularly confirmed to have PWS (Table 2b, cases 9 and 10; Table 2c, cases 10 and 11). Therefore, the size of ROH on chromosome 15 cannot be used as a reliable predictor of whether the patient had UPD15 or not. This underlines the importance of follow-up analysis for these patients even if the ROH may not be large. The HMZ segments in cases with mixed iso- and heteroUPD did not show a significantly greater concentration of HMZ segments at the subtelomeric or terminal regions. This is somewhat surprising and contrary to theoretical expectations that single-chromosome ROHs reflective of UPD would be more frequent at subtelomeric regions and signify true illegitimate meiotic recombination.4 There does not seem to be an easy explanation for this somewhat discrepant observation and our findings need further support from additional large studies. Future studies complemented by molecular confirmation of suspected UPD cases will be valuable. As an example, a case with 13 Mb of ROH (case 10, Table 2c) was located at 15q26.1-q26.3 (chr15:89 356 831–102 165 080) that is telomeric; another with 16 Mb of ROH (case 11, Table 2c) at 15q15.1-q21.3 (chr15:42 413 212–58 822 785) was not telomeric. We also demonstrated that parental origin of UPD15 can be identified by SNP array genotyping analysis if parental array data are available (Table 2c, case 8). The proband had two regions of homozygosity from chromosome 15 (a total of 47 Mb). A comparison of the genotype data between the proband and the mother disclosed both of the chromosomes 15 were inherited from the mother (UPD15mat). Molecular testing confirmed the diagnosis of PWS. Therefore, availability of parental genotype data allows imputation of parent of origin and may avoid the need for additional molecular studies in some cases.
For other chromosomes, it is likely that a significant number of these cases with ROH <25% of the whole chromosome in size and restricted to a single chromosome (although raising the possibility of segmental or whole chromosome UPD) may be coincidental in nature and molecular testing may exclude UPD (for example, case 13 in Table 2c). However, until a substantial number of such cases are actually evaluated by molecular analysis, no conclusion may be drawn.
Cytogenetic rearrangements have been widely known to cause UPD.13 The concurrent presence of marker chromosome or mosaic trisomy and single-chromosomal ROH aids in the diagnosis of UPD.13, 14 Our finding also strongly supports the use of oligo-SNP arrays in the evaluation of marker chromosomes (even when mosaic) and mosaic trisomy as it may provide crucial evidence not only regarding the identity of the marker chromosome and trisomy, but also help evaluate the possibility of UPD for the chromosome of origin. In the same context, G-banded chromosome and FISH analysis may prove beneficial in cases with UPD by identifying or ruling out the presence of small marker chromosomes or mosaic trisomy. Furthermore, as shown in our study, a genotype-first approach to identification of recessive disorders is facilitated by identifying ROHs particularly in consanguineous families and those with increased risk for recessive Mendelian disorders. Selection of candidate genes for specific autosomal recessive disorders from the regions with allelic homozygosity, followed by sequencing analysis to unravel the homozygous mutations, reflects significant benefits of SNP array analysis in solving the puzzle for specific phenotypes.1 Although it is challenging to limit the list of candidate disease genes to be considered in cases with extensive ROHs, analytical tools such as that developed by Wierenga et al12 has proven invaluable especially when phenotypic information can be used to limit the genes/disorder to be considered. With the additional aid of whole genome or exon sequencing, this type of analysis may become the principal tool to finding molecular causes for disorders in patients with regions of allelic homozygosity.
References
Sund KL, Zimmerman SL, Thomas C et al: Regions of homozygosity identified by SNP microarray analysis aid in the diagnosis of autosomal recessive disease and incidentally detect parental blood relationships. Genet Med 2013; 15: 70–78.
Kearney HM, Kearney JB, Conlin LK : Diagnostic implications of excessive homozygosity detected by SNP-based microarrays: consanguinity, uniparental disomy, and recessive single-gene mutations. Clin Lab Med 2011; 31: 595–613, ix.
Gibson J, Morton NE, Collins A : Extended tracts of homozygosity in outbred human populations. Hum Mol Genet 2006; 15: 789–795.
Papenhausen P, Schwartz S, Risheg H et al: UPD detection using homozygosity profiling with a SNP genotyping microarray. Am J Med Genet A 2011; 155A: 757–768.
Sasaki K, Mishima H, Miura K, Yoshiura K : Uniparental disomy analysis in trios using genome-wide SNP array and whole-genome sequencing data imply segmental uniparental isodisomy in general populations. Gene 2013; 512: 267–274.
McQuillan R, Leutenegger AL, Abdel-Rahman R et al: Runs of homozygosity in European populations. Am J Hum Genet 2008; 83: 359–372.
Campbell H, Carothers AD, Rudan I et al: Effects of genome-wide heterozygosity on a range of biomedically relevant human quantitative traits. Hum Mol Genet 2007; 16: 233–241.
Rudan I, Smolej-Narancic N, Campbell H et al: Inbreeding and the genetic complexity of human hypertension. Genetics 2003; 163: 1011–1021.
Ismail J, Jafar TH, Jafary FH, White F, Faruqui AM, Chaturvedi N : Risk factors for non-fatal myocardial infarction in young South Asian adults. Heart (British Cardiac Society) 2004; 90: 259–263.
Grote L, Myers M, Lovell A, Saal H, Lipscomb Sund K : Variability in laboratory reporting practices for regions of homozygosity indicating parental relatedness as identified by SNP microarray testing. Genet Med 2012; 14: 971–976.
Rehder CW, David KL, Hirsch B, Toriello HV, Wilson CM, Kearney HM : American College of Medical Genetics and Genomics: standards and guidelines for documenting suspected consanguinity as an incidental finding of genomic testing. Genet Med 2013; 15: 150–152.
Wierenga KJ, Jiang Z, Yang AC, Mulvihill JJ, Tsinoremas NF : A clinical evaluation tool for SNP arrays, especially for autosomal recessive conditions in offspring of consanguineous parents. Genet Med 2013; 15: 354–360.
Liehr T : Cytogenetic contribution to uniparental disomy (UPD). Mol Cytogenet 2010; 3: 8.
Liehr T, Ewers E, Hamid AB et al: Small supernumerary marker chromosomes and uniparental disomy have a story to tell. J Histochem Cytochem 2011; 59: 842–848.
Gao HZ, Kobayashi K, Tabata A et al: Identification of 16 novel mutations in the argininosuccinate synthetase gene and genotype-phenotype correlation in 38 classical citrullinemia patients. Hum Mutat 2003; 22: 24–34.
Engel K, Hohne W, Haberle J : Mutations and polymorphisms in the human argininosuccinate synthetase (ASS1) gene. Hum Mutat 2009; 30: 300–307.
Kirin M, McQuillan R, Franklin CS, Campbell H, McKeigue PM, Wilson JF : Genomic runs of homozygosity record population history and consanguinity. PLoS One 2010; 5: e13996.
Grote L, Myers M, Lovell A, Saal H, Sund KL : Variable approaches to genetic counseling for microarray regions of homozygosity associated with parental relatedness. Am J Med Genet A 2013; 164A: 87–98.
Schaaf CP, Scott DA, Wiszniewska J, Beaudet AL : Identification of incestuous parental relationships by SNP-based DNA microarrays. Lancet 2011; 377: 555–556.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Wang, JC., Ross, L., Mahon, L. et al. Regions of homozygosity identified by oligonucleotide SNP arrays: evaluating the incidence and clinical utility. Eur J Hum Genet 23, 663–671 (2015). https://doi.org/10.1038/ejhg.2014.153
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.153
This article is cited by
-
Chromosomal microarray analysis for prenatal diagnosis of uniparental disomy: a retrospective study
Molecular Cytogenetics (2024)
-
A cohort study of neurodevelopmental disorders and/or congenital anomalies using high resolution chromosomal microarrays in southern Brazil highlighting the significance of ASD
Scientific Reports (2024)
-
Frequent copy number variants in a cohort of Mexican-Mestizo individuals
Molecular Cytogenetics (2023)
-
Chromosomal microarray analyses from 5778 patients with neurodevelopmental disorders and congenital anomalies in Brazil
Scientific Reports (2022)
-
Diagnostic yield of additional exome sequencing after the detection of long continuous stretches of homozygosity (LCSH) in SNP arrays
Journal of Human Genetics (2021)