Abstract
Holt–Oram syndrome (HOS) is a rare autosomal dominant heart–hand syndrome due to mutations in the TBX5 transcription factor. Affected individuals can have structural cardiac defects and/or conduction abnormalities, and exclusively upper limb defects (typically bilateral, asymmetrical radial ray defects). TBX5 mutations reported include nonsense, missense, splicing mutations and exon deletions. Most result in a null allele and haploinsufficiency, but some impair nuclear localisation of TBX5 protein or disrupt its interaction with co-factors and downstream targets. We present a five generation family of nine affected individuals with an atypical HOS phenotype, consisting of ulnar ray defects (ulnar hypoplasia, short fifth fingers with clinodactyly) and very mild radial ray defects (short thumbs, bowing of the radius and dislocation of the radial head). The cardiac defects seen are those more rarely reported in HOS (atrioventricular septal defect, hypoplastic left heart syndrome, mitral valve disease and pulmonary stenosis). Conduction abnormalities include atrial fibrillation, atrial flutter and sick sinus syndrome. TBX5 mutation screening (exons 3–10) identified no mutations. Array comparative genomic hybridisation (CGH) revealed a 48 kb duplication at 12q24.21, encompassing exons 2–9 of the TBX5 gene, with breakpoints within introns 1–2 and 9–10. The duplication segregates with the phenotype in the family, and is likely to be pathogenic. This is the first known report of an intragenic duplication of TBX5 and its clinical effects; an atypical HOS phenotype. Further functional studies are needed to establish the effects of the duplication and pathogenic mechanism. All typical/atypical HOS cases should be screened for TBX5 exon duplications.
Similar content being viewed by others
Introduction
Holt–Oram syndrome (HOS; MIM 142900) affects ∼1 in 100 000 livebirths and is one of many hand–heart syndromes. It was first described as a syndrome of upper limb abnormalities and congenital heart defects by Holt and Oram.1 It is inherited in an autosomal dominant manner with high penetrance and variable expression (ranging from only subclinical skeletal findings to phocomelia, and from conduction defects to severe congenital heart defects). The typical combination is thought to be triphalangeal thumbs and a secundum atrial septal defect (ASD), but there is a range in severity of limb and heart defects. These individuals do not have problems with growth or development.2 There is no specific facial profile that discriminates it from other hand–heart syndromes.3
Upper limb abnormalities are always present and most commonly bilateral and asymmetrical (left side more than right).4, 5 They involve structures derived from the radial ray (radius, carpal and thenar bones), which can show aplasia, hypoplasia, fusion or anomalous development. This results in features ranging from limitation in movement, triphalangeal or absent thumbs, foreshortened forearms, to phocomelia.1, 4 The thumbs are the most commonly affected structure (84%), with radial aplasia/hypoplasia in 64%. Ulnar ray defects have been shown to occur only when the radial ray is involved, and to a lesser degree of severity.5 Smith et al4 showed that the radius was affected in 22/39 cases, but the ulna was only involved in 15/39. The humerus was hypoplastic in 50% cases, and the clavicle was sometimes abnormal. Many show a characteristic appearance of narrow, sloping shoulders, because of a combination of short clavicles, hypoplasia of head of humerus and decreased musculature. Poznanski et al6 demonstrated that carpal abnormalities are more specific to HOS than changes to the thumbs, particularly the scaphoid bone. The lower limbs are usually normal.
Heart defects typically are septal defect, more commonly ASDs or rarely ventricular septal defects (VSDs), but other more severe heart defects have been described (eg, tetralogy of Fallot, truncus arteriosus and hypoplastic left heart syndrome (HLHS)).5, 7 In familial cases of HOS, 95% affected individuals have been shown to have a heart defect. Cardiac conduction abnormalities are commonly found on an electrocardiogram (ECG), and include atrioventricular block, bradycardia and atrial fibrillation (AF). They are more likely to occur in those with a structural heart defect, but 40% cases have conduction abnormalities alone without heart defects.
There is not thought to be any association between the severity of limb and heart defects. Penetrance is thought to be close to 100% (if investigated by a wrist X-ray and ECG/ECHO). Around 60–70% of patients have an affected parent, whereas 30–40% cases are de novo.8 Basson et al9 and Terrett et al10 showed linkage of the disease to 12q2 via familial genetic analysis. The candidate gene was identified as TBX5 (MIM 601620), a member of the Brachyury(T) gene family, which acts as a transcription factor and the conserved T-box domain serves as a DNA-binding domain. Molecular analysis of HOS familial and sporadic cases demonstrated mutations within this gene.11, 12 Most of the mutations result in a null allele and haploinsufficiency, but some have been shown to impair the nuclear localisation of TBX5 protein or disrupt TBX5 interaction with co-factors and downstream target genes.13, 14
We describe a five generation family with an atypical HOS phenotype, where nine affected individuals have combinations of (a) mild limb defects affecting mainly the ulnar ray, (b) non-ASD/VSD heart defects and/or (c) rhythm anomalies. We identified a novel 48 kb intragenic duplication in TBX5, which co-segregates with the disease phenotype. This is the first report of a duplication within TBX5 as the mutation type causing HOS, and is likely to result in the atypical HOS phenotype in this family.
Clinical report
Consultand
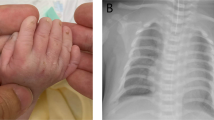
The consultand (IV:3) (23-year-old female) is the first child of non-consanguineous White British parents, who was referred to the Clinical Genetics department for an assessment to identify a possible diagnosis. She was born with an atrioventricular septal defect (AVSD) and pulmonary stenosis (PS), which had been repaired surgically, and reported to have congenital bilateral upper limb defects. She subsequently developed atrial flutter in her teens, which was pharmacologically controlled. There were no other significant details from her perinatal or developmental history. On clinical examination, the findings were short fifth fingers bilaterally (particularly the terminal phalanx), 2/3 syndactly, mild deformity of right index finger, thenar hypoplasia, and bilateral short, broad thumbs (Figure 1a). She had short forearms with difficulty in forearm supination and pronation, and a prominence on both of her elbows representing subluxation of the radial heads. She had no facial dysmorphism and no other upper or lower limb defects. X-rays of her hands confirmed the above clinical findings, but also identified bilateral distal ulnar hypoplasia (leading to bowing of the radii and dislocation of the radial heads) and bilateral abnormal trochlear notches at the proximal ulna (leading to abnormal humero-ulnar joints) (Figures 2a and b). The carpal bones were reported as being entirely normal. Previously, routine cytogenetic analysis revealed a normal female karyotype, and bidirectional sequencing of coding exons 3–10 of the TBX5 gene did not identify any mutations or variants.
Photographs of both hands of affected family members. (a) IV:3 (Consultand) – short fifth fingers especially terminal phalanx, short thumbs, 2/3 syndactyly, and mild deformity of the right index finger. (b) IV:5 (Sister of consultand) – short fifth fingers and short thumbs. (c) III:5 (Mother of consultand) – short fifth fingers, fifth finger clinodactyly, and short thumbs. (d) III:3 (Maternal aunt of consultand) – short fifth fingers, fifth finger clinodactyly, and slightly abnormal thumbs. All four individuals above also had thenar hypoplasia (not shown in figure images).
Family members
The pedigree shows 9/12 individuals (including the consultand) over five generations within the family, with features suggestive of HOS and a clear autosomal dominant inheritance pattern (Figure 3). Clinical examination was not possible in four deceased individuals (thought to be affected) but clinical details were obtained. After the consultand, all remaining family members (7/11) were assessed clinically (7/11) and radiologically (1/11), and the findings are summarised in Table 1. The consistent limb defects in affected individuals include: short fifth fingers (6/9), fifth finger clinodactyly (3/9), short thumbs (4/9), thenar hypoplasia (5/9) and difficulty in forearm supination and pronation (4/9) (Figures 1b–d). X-rays of one other affected individual (III:3) were reported as normal. There were no individuals identified/reported with severe radial ray defects (absent/triphalangeal thumbs or radial aplasia/hypoplasia). Cardiac defects present in 5/9 affected individuals include: AVSD with PS (1/5), HLHS (1/5), mixed mitral valve disease (2/5) and a septal defect (1/5). AF (2/5), atrial flutter (1/5), sick sinus syndrome (1/5) and unknown rhythm disturbance (1/5) were the conduction abnormalities identified/reported in the family (5/9), mainly in the absence of a cardiac defect (3/5), however, 3/4 remaining individuals with normal ECG parameters had cardiac defects.
Materials and methods
Array studies
Genomic DNA was extracted from peripheral blood using the AutoPure extraction system (Qiagen, Crawley, UK) following the manufacturer's instructions. DNA concentration and purity (A260−280∼1.8–2.0) was determined using a NanoDrop spectrophotometer (Labtech International, Ringmer, UK). Samples were labelled with Cy3 and Cy5 using the BlueGnome Fluorescent Labelling System (dUTP/oligo; BlueGnome, Cambridge, UK) according to manufacturer's instructions. In all, 500 ng genomic DNAs were hybridised to BlueGnome 8 × 60k International Standard Cytogenomic Array design oligonucleotide microarray and arrays washed post-hybridisation according to the manufacturer's instructions. Arrays were scanned using an Agilent C scanner (G2939A) and analysed in BlueFuse Multi v2.2 (BlueGnome). This platform should detect the majority of copy number imbalances >15 kb in 500 disease gene/telomeric regions and >180 kb in the genomic backbone and may detect smaller imbalances in some instances. Oligonucleotides were mapped to genome build NCBI36. Genomic locations were later converted to NCBI genome build GRCh37 using NCBI's remapping tool.
Multiplex ligation-dependent probe amplification
Multiplex ligation-dependent probe amplification (MLPA) analysis of the TBX5 gene was carried out using the MRC-Holland P180-B1 kit (MRC Holland, Amsterdam, The Netherlands) according to the manufacturer's instructions and using a Tetrad PCR machine (BioRad, Hercules, CA, USA). Capillary electrophoresis was carried out on an ABI 3130 (Applied Biosystems, Durmstadt, Germany) and analysed using the Soft Genetics Gene Marker v1.70 program (Softgenetics LLC, State College, PA, USA). This technique will detect copy number changes in all 10 exons of TBX5 (NCBI GRCh37) (Genebank accession number NM_000192.3, exon 2 is only present in transcript NM_181486.1).
Results
Identification of duplication on chromosome 12 using array-comparative genomic hybridisation (CGH)
Array analysis showed an approximately 48 kb duplication at 12q24.21. The duplicated region contained 17 oligonucleotide probes, genomic co-ordinates 12:114795705–114844082 (NCBI GRCh37) (Figure 4). The breakpoints of the duplication occur within introns 9–10 (proximally) and introns 1–2 (distally) of the TBX5 gene, implying exons 2–9 are duplicated and exons 1 and 10 are intact. Two other affected family members (III:3 and V:1) analysed using array-CGH were identified to have the same duplication.
Comparison of the copy number imbalance within TBX5 using bioinformatic tools
Comparison of the duplicated region in this case to other copy number imbalances involving the TBX5 gene was carried out using publicly available web-based bioinformatic tools (DECIPHER, Database of Genomic Variants (DGV), Ensembl, HGNC, OMIM and Pubmed). There is no recorded benign copy number variation in the DGV and to our knowledge no reports of a duplication within TBX5 in the literature to date.
Confirmation of duplication of the TBX5 gene by MLPA
Analysis of the TBX5 gene using MLPA in other family members (7/11) showed a heterozygous duplication of exons 2–9 of TBX5 segregating with clinically affected family members only, thereby confirming the array results (Table 2).
Discussion
This is a novel intragenic duplication in TBX5 present in a large family with an atypical form of HOS, where cardiac defects (structural and/or conduction) predominate, and the limb defects are mild. The five generation family with nine known/suspected individuals with HOS shows marked intrafamilial variability with cardiac involvement in 8/9 (89%), varying from isolated conduction abnormalities like AF to more complex heart defects like HLHS. Non-septal cardiac defects predominate in this family (all of which have been reported in previous cases of HOS, but not commonly).5, 7 The classical conduction abnormalities of heart block and bradycardia were not present in the family, but AF was present instead. Only 2/9 (22%) individuals had both structural heart defects and conduction abnormalities; structural cardiac defects in absence of conduction abnormalities were present in 3/9 (33%); and conduction abnormalities in absence of structural cardiac defects were present in 3/9 (33%). The commonly observed findings of absent/triphalangeal thumbs (62%) and/or aplastic/hypoplastic radii (64%) were not seen in any of the affected individuals in our family. One may usually expect ulnar ray anomalies to only occur alongside radial ray ones. Our family showed ulnar ray anomalies, sometimes in the absence of any radial ray ones (III:3). The consistent limb findings within the family were short fifth fingers (66%), fifth finger clinodactyly (44%), short thumbs (44%), thenar hypoplasia (56%) and limitations in forearm supination/pronation (44%). These features were reported to occur in 43%, 64%, 39%, 70% and 61% of cases, respectively, in a large-scale clinical study of HOS cases.5
Several families have been reported to show anticipation in successive generations with no obvious genetic mechanism and explanation,5, 15, 16, 17 and this phenomenon could be observed in the cardiac features in our family. Generations I–III had predominantly conduction abnormalities, and there is the occurrence of increasing severity of congenital heart defects in generations III–V (III:1-septal defect; IV:3-AVSD/PS; V:1-HLHS), but this could, however, be ascertainment bias. A maternal effect has been described with greater severity seen in offspring of affected females,5 and although there are not many males in our family, the offspring of some females have a more severe phenotype implying a possible maternal effect (mother (III:3) with AF compared with her daughter (IV:3) with AVSD/PS and atrial flutter; mother (IV:5) with no cardiac defects compared with her daughter (V:1) with HLHS).
Suspected cases of HOS (and both their parents) should have a thorough examination to detect abnormalities either in shape/size/function of the thumb. The forearms should be assessed for ability to pronate and supinate. Radiology is likely to help in clinically mildly affected individuals. Cross et al13 showed only 30% of cases with a clinical diagnosis of HOS had a TBX5 mutation, but this low detection rate can be markedly improved when coupled with adherence to strict diagnostic criteria (ie, personal and/or /family history of cardiac septation and/or conduction defects in the setting of preaxial radial ray deformity).18 Those who have a suspected diagnosis of HOS (but do not meet the diagnostic criteria) and do not have a TBX5 mutation often have clinical features that overlap with HOS but also have other notable atypical clinical manifestations (eg, ulnar or lower limb involvement, syndactyly of digits other than the thumb, polydactyly or craniofacial findings). Ulnar-mammary syndrome (due to TBX3 gene mutations) is one of the differential diagnoses to be considered as it involves cardiac defects similar to HOS and ulnar ray defects.
The diagnosis of HOS has always been suspected in our family as they meet the diagnostic criteria, despite milder limb defects mainly affecting the ulnar side. Therefore, genetic counselling in this family has been difficult over the years as no mutations were identified in TBX5. Sletten et al7 highlighted the fact that severe heart defects in patients with HOS have been underestimated and should be considered in counselling families with HOS. The cardiac phenotype seems to predominate in this family with the unfortunate recent death of a child with HLHS (V:1). This makes it essential that the family are counselled on the variability of this condition, as there may be a risk of complex heart defects in future offspring/generations. The family could also be at risk of more severe limb defects despite affected individuals having a milder limb phenotype, and no individuals with radial aplasia/hypoplasia. The identification of a TBX5 duplication, which segregates with the phenotype in the family is likely to explain the HOS features in this family. The family can now be offered prenatal testing to clarify risks to a pregnancy, and genetic testing to clinically unaffected individuals for clarification of their status. This was performed via MLPA in three clinically unaffected individuals in the family (III:3, IV:1 and IV:2), which also helped to clarify segregation of the duplication with the disease phenotype.
The T-box gene family consists of a large family of transcription factors that have diverse roles in embryonic development and a number of human disorders have been linked to mutations in these genes (TBX5/HOS; TBX3/ulnar-mammary syndrome; and TBX1/DiGeorge syndrome).19 The coding region of TBX5 is exons 3–10 and exons 1 and 2 are untranslated but have been shown to be critical for normal TBX5 expression.20, 21 TBX5 expression is documented in the early development of the heart in mice, chicks and humans and of the forelimb.11, 12, 22
Most TBX5 mutations are nonsense mutations resulting in a truncated TBX5 protein and haploinsufficiency (via nonsense-mediated mRNA degradation).21, 23, 24 Deletions of the whole gene or exons are thought to cause abnormalities equivalent to those mutations causing a truncated protein, because of reduced TBX5 dosage.25, 26 A contiguous deletion of 2.19–2.27 Mb encompassing the TBX5 and TBX3 genes was identified in one family with clinical features of HOS and ulnar-mammary syndrome.27
The pathogenetic mechanism by which protein producing mutations (missense mutations and in frame deletions) would cause haploinsufficiency is not well understood as they would not alter TBX5 dosage. The normal cellular localisation of the TBX5 protein is in the nucleus in cardiomyocytes.28 Fan et al29, 30 analysed various missense mutations and in frame deletions and described their possible pathogenetic mechanisms as either: (a) abnormal cellular localisation of mutant TBX5 protein or (b) altered interactions with cardiac or limb specific co-factors (eg, NKX2.5, GATA4, Cx40 and SALL4). They showed increased concentrations within the cytoplasm of TBX5 mutant proteins from various missense mutations and one in-frame deletion, providing evidence of altered nuclear localisation. They also showed the Gly80Arg, Arg237Gln and Arg237Trp mutations dramatically reduce the DNA-binding ability of TBX5, and result in reduced transcriptional activation of TBX5-dependent genes and loss of synergistic transcriptional activation with NKX2.5. TBX5 is known to associate with NKX2.5 and synergistically promote cardiomyocyte differentiation.31 Other specific missense mutations in TBX5 have been shown to disrupt the interaction between TBX5 and GATA4 (a gene known to cause non-syndromic congenital heart disease, particularly septal defects). Therefore, mutations in GATA4 are likely to cause septal defects through its interaction with TBX5.32 Mouse models of HOS (TBX5del/+heterozygote) show dramatic reduced expression of Cx40 (connexion 40), a gene important for the conduction of electrical impulses throughout the heart, and therefore accounting for the conduction defects seen in HOS.33 Böhm et al34 described a deletion mutation, which resulted in an elongated TBX5 protein that showed normal cellular localisation but reduced activation of a downstream target – atrial natriuretic factor. Mutations in SALL4 (associated with Duane-radial ray syndrome) cause similar upper limb and heart defects as mutations in TBX5. Koshiba-Takeuchi et al35 showed Tbx5 and Sall4 interact both positively and negatively to finely regulate patterning and morphogenesis of the anterior forelimb and heart in mice, providing a mechanism for these hand–heart syndromes.
Genotype–phenotype correlations have been proposed by some groups. Mutations creating null alleles cause more severe abnormalities in both limb and heart development.21 Some missense mutations in TBX5 can also produce distinct phenotypes, such that those at the 5′ end of the gene (eg, Gly80Arg) more significantly lead to cardiac defects and only minor limb defects, whereas those at the 3′ end (eg, Arg237Gln and Arg237Trp) more significantly cause limb defects and only minor cardiac defects.21 A broader study by Brassington et al36 found no evidence to support the notion that the type or location of mutation was predictive of severity of clinical features. These studies do not confirm or rule out the possibility of these genotype–phenotype relationships, as they all involve small numbers of cases, inadequate for any statistical analyses.
Interestingly some studies have shown that the cellular biological consequences of both diminished and augmented Tbx5 expression are similar, particularly in relation to cardiac development.37, 38, 39, 40 Patients with whole or partial chromosome 12q duplications (including the 12q24.21 region and TBX5 gene) have been described to have some of the cardiac features of HOS (mainly septal defects), other congenital anomalies and dysmorphism, but hardly any of the skeletal features.41, 42, 43, 44, 45, 46, 47, 48, 49 This partially conforms to the prominent cardiac defects and milder limb defects seen in this family with the TBX5 duplication, therefore it seems expression of TBX5 must be finely regulated for normal cardiac and limb development. Postma et al50 described a HOS family with predominantly cardiac defects and mild limb defects, with a novel gain-of-function missense mutation (Gly125Arg) in TBX5. The TBX5 mutant protein showed a normal NKX2.5 interaction, normal nuclear localisation, but had significantly enhanced DNA-binding properties and increased expression of many downstream target genes (Cx40, Tbx3, Nppa and Kcnj2). McDermott et al51 commented that HOS patients can exhibit paroxysmal AF on ambulatory ECG recordings and this can be in the absence of congenital structural heart defects. They stress that clinicians should not predict classes of mutations based on the clinical phenotype because of marked intra- and interfamilial variability.
It is likely that the duplication identified in our family is in tandem, but this is difficult to prove as localisation studies with FISH are not possible due to the small size of the duplication. TBX5 is not expressed in lymphocytes so further analysis on cardiomyocytes will help to clarify the exact pathogenetic mechanism. The Tbx2, Tbx3, Tbx4 and Tbx5 genes arose by duplication events of an ancestral Tbx2/34/5 locus. It is suggested essential cis-regulatory elements (located in flaking or intragenic regions) are likely to be found in close vicinity to these loci.52 Minguillon et al53 showed that changes at the regulatory level of Tbx4 and Tbx5 expression were necessary for the development of paired appendages during vertebrate evolution, rather than the generation of novel protein function. We speculate that the atypical HOS phenotype seen in this family could be due to a number of possible underlying pathogenetic mechanisms. The intragenic duplication could be in frame and cause its effects via: (a) an elongated protein leading to a gain-of-function, (b) an elongated protein with abnormal cellular localisation mimicking functional haploinsufficiency or (c) increased TBX5 expression (possibly via an intragenic cis-regulating element). However, we are unable to rule out an out of frame duplication causing its effects via a truncated protein, which undergoes nonsense-mediated mRNA degradation. As seen in the 12q duplication cases reported previously, the duplication in this family may explain the prominent cardiac defects and milder limb defects.
Conclusion
This family is the first reported case of an intragenic duplication in TBX5, which is associated with atypical HOS clinical features, and extends our molecular knowledge of TBX5 gene mutations. Further studies are required to determine the exact pathogenetic mechanism resulting from the duplication. This case highlights the value of array CGH in patients with an abnormal phenotype, and it is proposed that comprehensive testing for atypical and typical HOS should include analysis and reporting for copy number changes of TBX5 (including exon deletions and duplications).
Accession codes
References
Holt M, Oram S : Familial heart disease with skeletal malformations. Br Heart J 1960; 22: 236–242.
Hurst JA, Hall CM, Baraitser M : The Holt-Oram syndrome. J Med Genet 1991; 28: 406–410.
Allanson JE, Newbury-Ecob RA : Holt-Oram syndrome: is there a ‘face’? Am J Med Genet 2003; 118A: 314–318.
Smith AT, Sack Jr GH, Taylor GJ : Holt-Oram syndrome. J Pediatr 1979; 95: 538–543.
Newbury-Ecob RA, Leanage R, Raeburn JA, Young ID : Holt-Oram syndrome: a clinical genetic study. J Med Genet 1996; 33: 300–307.
Poznanski AK, Gall Jr JC, Stern AM : Skeletal manifestations of the Holt-Oram syndrome. Radiology 1970; 94: 45–53.
Sletten LJ, Pierpont ME : Variation in severity of cardiac disease in Holt-Oram syndrome. Am J Med Genet 1996; 65: 128–132.
Huang T : Current advances in Holt-Oram syndrome. Curr Opin Pediatr 2002; 14: 691–695.
Basson CT, Cowley GS, Solomon SD et al: The clinical and genetic spectrum of the Holt-Oram syndrome (heart-hand syndrome). N Engl J Med 1994; 330: 885–891.
Terrett JA, Newbury-Ecob R, Cross GS et al: Holt-Oram syndrome is a genetically heterogeneous disease with one locus mapping to human chromosome 12q. Nat Genet 1994; 6: 401–404.
Basson CT, Bachinsky DR, Lin RC et al: Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 1997; 15: 30–35.
Li QY, Newbury-Ecob RA, Terrett JA et al: Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet 1997; 15: 21–29.
Cross SJ, Ching YH, Li QY et al: The mutation spectrum in Holt-Oram syndrome. J Med Genet 2000; 37: 785–787.
Mori AD, Bruneau BG : TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr Opin Cardiol 2004; 19: 211–215.
McKusick VA : Genetic factors in cardiovascular diseases. J Am Geriatr Soc 1961; 9: 465–476.
Temtamy SA, McKusick VA : The genetics of hand malformations. Birth Defects Orig Artic Ser 1978; 14: i–xviii 1-619.
Gardner RJ, Buckfield PM, Veale AM : A girl with severe expression of the holt-oram gene. Clin Genet 1973; 4: 417–421.
McDermott DA, Bressan MC, He J et al: TBX5 genetic testing validates strict clinical criteria for Holt-Oram syndrome. Pediatr Res 2005; 58: 981–986.
Packham EA : T-box genes in human disorders. Human Mol Genet 2003; 12: 37R–344.
Yi CH, Russ A, Brook JD : Virtual cloning and physical mapping of a human T-box gene, TBX4. Genomics 2000; 67: 92–95.
Basson CT, Huang T, Lin RC et al: Different TBX5 interactions in heart and limb defined by Holt-Oram syndrome mutations. Proc Natl Acad Sci USA 1999; 96: 2919–2924.
Ghosh TK, Song FF, Packham EA et al: Physical interaction between TBX5 and MEF2C is required for early heart development. Mol Cell Biol 2009; 29: 2205–2218.
Yang J, Hu D, Xia J et al: Three novel TBX5 mutations in Chinese patients with Holt-Oram syndrome. Am J Med Genet 2000; 92: 237–240.
Gruenauer-Kloevekorn C, Froster UG : Holt-Oram syndrome: a new mutation in the TBX5 gene in two unrelated families. Annales de Génétique 2003; 46: 19–23.
Akrami SM, Winter RM, Brook JD, Armour JA : Detection of a large TBX5 deletion in a family with Holt-Oram syndrome. J Med Genet 2001; 38: E44.
Borozdin W, Bravo Ferrer Acosta AM, Bamshad MJ et al: Expanding the spectrum ofTBX5 mutations in Holt-Oram syndrome: detection of two intragenic deletions by quantitative real time PCR, and report of eight novel point mutations. Human Mutation 2006; 27: 975–976.
Borozdin W, Bravo-Ferrer Acosta AM, Seemanova E et al: Contiguous hemizygous deletion ofTBX5,TBX3, andRBM19 resulting in a combined phenotype of Holt-Oram and ulnar-mammary syndromes. Am J Med Genet Part A 2006; 140A: 1880–1886.
Hatcher CJ, Goldstein MM, Mah CS, Delia CS, Basson CT : Identification and localization of TBX5 transcription factor during human cardiac morphogenesis. Dev Dyn 2000; 219: 90–95.
Fan C, Duhagon MA, Oberti C et al: Novel TBX5 mutations and molecular mechanism for Holt-Oram syndrome. J Med Genet 2003; 40: e29.
Fan C, Liu M, Wang Q : Functional analysis of TBX5 missense mutations associated with Holt-Oram syndrome. J Biol Chem 2003; 278: 8780–8785.
Hiroi Y, Kudoh S, Monzen K et al: Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet 2001; 28: 276–280.
Garg V, Kathiriya IS, Barnes R et al: GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003; 424: 443–447.
Bruneau BG, Nemer G, Schmitt JP et al: A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001; 106: 709–721.
Böhm J, Heinritz W, Craig A et al: Functional analysis of the novel TBX5 c.1333delC mutation resulting in an extended TBX5 protein. BMC Med Genet 2008; 9: 88.
Koshiba-Takeuchi K, Takeuchi JK, Arruda EP et al: Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat Genet 2006; 38: 175–183.
Brassington AM, Sung SS, Toydemir RM et al: Expressivity of Holt-Oram syndrome is not predicted by TBX5 genotype. Am J Hum Genet 2003; 73: 74–85.
Hatcher CJ, Kim MS, Basson CT : Atrial form and function: lessons from human molecular genetics. Trends Cardiovasc Med 2000; 10: 93–101.
Liberatore CM, Searcy-Schrick RD, Yutzey KE : Ventricular expression of tbx5 inhibits normal heart chamber development. Dev Biol 2000; 223: 169–180.
Hatcher CJ, Kim MS, Mah CS et al: TBX5 transcription factor regulates cell proliferation during cardiogenesis. Dev Biol 2001; 230: 177–188.
Hatcher CJ, Diman NY, Kim MS et al: A role for Tbx5 in proepicardial cell migration during cardiogenesis. Physiol Genomics 2004; 18: 129–140.
Roberts SH, Mattina T, Laurence KM, Sorge G, Pavone L : Partial trisomy 12q: report of a case and review. J Med Genet 1981; 18: 470–473.
Melnyk AR, Weiss L, Van Dyke DL, Jarvi P : Malformation syndrome of duplication 12q24.1 leads to qter. Am J Med Genet 1981; 10: 357–365.
Pratt NR, Bulugahapitiya DT : Partial trisomy 12q: a clinically recognisable syndrome. Genetic risks associated with translocations of chromosome 12q. J Med Genet 1983; 20: 86–89.
Tengström C, Wilska M, Kähkönen M, Autio S, Leisti J : Partial trisomy 12q: clinical and cytogenetic observations. Clin Genet 1985; 28: 112–117.
McCorquodale MM, Rolf J, Ruppert ES, Kurczynski TW, Kolacki P : Duplication (12q) syndrome in female cousins, resulting from maternal (11;12) (p15.5;q24.2) translocations. Am J Med Genet 1986; 24: 613–622.
Dixon JW, Costa T, Teshima IE : Mosaicism for duplication 12q (12q13q24.2) in a dysmorphic male infant. J Med Genet 1993; 30: 70–72.
Vaughan CJ, Basson CT : Molecular determinants of atrial and ventricular septal defects and patent ductus arteriosus. Am J Med Genet 2000; 97: 304–309.
Doco-Fenzy M, Mauran P, Lebrun JM et al: Pure direct duplication (12)(q24.1 → q24.2) in a child with Marcus Gunn phenomenon and multiple congenital anomalies. Am J Med Genet Pt A 2006; 140A: 212–221.
Shchelochkov OA, Patel A, Weissenberger GM et al: Duplication of chromosome band 12q24.11q24.23 results in apparent Noonan syndrome. Am J Med Genet Pt A 2008; 146A: 1042–1048.
Postma AV, van de Meerakker JB, Mathijssen IB et al: A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circulation Res 2008; 102: 1433–1442.
McDermott DA, Hatcher CJ, Basson CT : Atrial fibrillation and other clinical manifestations of altered TBX5 dosage in typical Holt-Oram syndrome. Circulation Res 2008; 103: e96–e96.
Horton AC, Mahadevan NR, Minguillon C et al: Conservation of linkage and evolution of developmental function within the Tbx2/3/4/5 subfamily of T-box genes: implications for the origin of vertebrate limbs. Dev Genes Evol 2008; 218: 613–628.
Minguillon C, Gibson-Brown JJ, Logan MP : Tbx4/5 gene duplication and the origin of vertebrate paired appendages. Proc Natl Acad Sci USA 2009; 106: 21726–21730.
Acknowledgements
We are particularly grateful for the family who supported our publication of their case to benefit other families.
Web resources
Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER), http://decipher.sanger.ac.uk
Database of Genomic Variants (DGV), http://projects.tcag.ca/variation
Ensembl, http://www.ensembl.org/index.html
HUGO Gene Nomenclature Committee (HGNC), http://www.genenames.org/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim
Pubmed, http://www.ncbi.nlm.nih.gov/pubmed/
NCBI genome remapping tool, http://www.ncbi.nlm.nih.gov/genome/tools/remap/
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Patel, C., Silcock, L., McMullan, D. et al. TBX5 intragenic duplication: a family with an atypical Holt–Oram syndrome phenotype. Eur J Hum Genet 20, 863–869 (2012). https://doi.org/10.1038/ejhg.2012.16
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.16
Keywords
This article is cited by
-
Radiographic assessment of congenital malformations of the upper extremity
Pediatric Radiology (2016)
-
Myocardial transcription factors in diastolic dysfunction: clues for model systems and disease
Heart Failure Reviews (2016)
-
Novel TBX5 Duplication in a Japanese Family with Holt–Oram Syndrome
Pediatric Cardiology (2015)
-
Holt Oram syndrome: a registry-based study in Europe
Orphanet Journal of Rare Diseases (2014)