Abstract
Y-chromosome microsatellites (Y-STRs) are typically used for kinship analysis and forensic identification, as well as for inferences on population history and evolution. All applications would greatly benefit from reliable locus-specific mutation rates, to improve forensic probability calculations and interpretations of diversity data. However, estimates of mutation rate from father–son transmissions are available for few loci and have large confidence intervals, because of the small number of meiosis usually observed. By contrast, population data exist for many more Y-STRs, holding unused information about their mutation rates. To incorporate single locus diversity information into Y-STR mutation rate estimation, we performed a meta-analysis using pedigree data for 80 loci and individual haplotypes for 110 loci, from 29 and 93 published studies, respectively. By means of logistic regression we found that relative genetic diversity, motif size and repeat structure explain the variance of observed rates of mutations from meiosis. This model allowed us to predict locus-specific mutation rates (mean predicted mutation rate 2.12 × 10−3, SD=1.58 × 10−3), including estimates for 30 loci lacking meiosis observations and 41 with a previous estimate of zero. These estimates are more accurate than meiosis-based estimates when a small number of meiosis is available. We argue that our methodological approach, by taking into account locus diversity, could be also adapted to estimate population or lineage-specific mutation rates. Such adjusted estimates would represent valuable information for selecting the most reliable markers for a wide range of applications.
Similar content being viewed by others
Introduction
Around 400 microsatellite markers from the Y human chromosome have been made available to date (eg, Hanson and Ballantyne1), with important applications in forensic analyses and in genealogy research. However, reliable locus-specific mutation rates are needed to carefully choose loci to minimize the error rate in kinship analysis and sample identification2 while obtaining the maximum discriminatory power (eg, Mulero et al, Lim et al and Vermeulen et al3, 4, 5). Also in population genetics and evolutionary studies, correct inferences on the timing of major demographic events, the age of the most common ancestor, as well as dating Y-lineages and tracing disease evolution are based on the knowledge of mutation rates (eg, Shi et al, Zerjal et al and Xue et al6, 7, 8).
Population genetic theory predicts the genetic diversity of loci in function of their mutation rates (μ) and the effective size of populations (N). Therefore, it is possible to obtain estimates of the joint parameter θ=2Nμ from genetic diversity indices. In the case of loci evolving under a stepwise mutation model (SMM, generally assumed for microsatellites) it is possible to use the variance (V) in allele repeat count (ie, allele size measured in number of repeats) and the ‘homozygosity’ ( , where k is the number of different alleles in the population and pi is the frequency of the ith allele; note that the term homozygosity is not biologically meaningful for haploid loci, but it will be used through the article for the sake of simplicity) for the estimation of θ using the following relationships:9
, where k is the number of different alleles in the population and pi is the frequency of the ith allele; note that the term homozygosity is not biologically meaningful for haploid loci, but it will be used through the article for the sake of simplicity) for the estimation of θ using the following relationships:9


where the hat denotes estimated values. However, because it is difficult to separate the effects of demography (ie, N), estimates of θ provide little information about mutation rate. Nevertheless, it is possible to obtain information about relative mutation rates. In the case of effective population size being the same among loci within one population (ie, neutral loci with same ploidy level, such as the Y-STRs), the ratio between the θ of two loci should be the same as the ratio between their mutation rates.10 However, relative mutation rate estimates have limited utility for dating evolutionary events or calculating forensic probabilities.
Absolute mutation rate estimates can be obtained by the analysis of allele transmissions in pedigrees (eg, Heyer et al and Gusmão et al11, 12). The proportion of allele mismatches in father–son transmissions is currently the most widely used approach to obtain estimates of mutation rates for Y-STRs. Owing to the low values of mutation rates, large number of father–son pairs must be genotyped to obtain accurate estimates. This has limited the number of Y-STR loci for which these estimates exist and many of them have been obtained from rather low sample sizes. On the other hand, population diversity data exist for many more Y-STRs, holding unused information about their mutation rates. The objective of this work is to present a method to combine pedigree and population data for the estimation of mutation rates and to provide locus-specific mutation rate estimates for 110 Y-STR loci (71 of which had no previous estimate).
Materials and methods
Source of population data
Population data for 110 Y-chromosome microsatellite loci have been collected from 93 published works, for a total of 22 165 individual haplotypes (note that each individual was genotyped for a subset of loci and never for all of them). Locus names, sample sizes and references are detailed in Supplementary Table S1. Locus nomenclature and allele call have been thoroughly checked to assure congruence across works and to remove duplicate data. Any population data with incongruent allele codes were either made uniform (when information provided by authors made it possible unequivocally) or excluded from analysis. Specifically, data from GATA H4 and GATA H4.1 have been pooled under the name GATA H4.1 by applying the appropriate correction to allele calls13, 14 and DYS389II has been transformed into DYS389B by subtracting allele size of DYS389I.15 Multicopy loci and single individuals with duplicated or variant alleles were excluded from the analysis. Data sets were chosen in order to obtain a maximum representation of loci and of geographical areas; collection of data stopped when no additional data sets could be found that would add data for new loci or would increase the order of magnitude of the sample size for individuals genotyped for a locus.
Source of meiosis (father–son pair) data
Direct observations of mutation events from meiosis data (father–son pairs) have been collected for 80 loci among the 110 loci with population data, from 29 published studies (Supplementary Table 1 and Supplementary Table S2). Confidence intervals (CIs) from binomial probability distribution were estimated according to Wilson method.16 Mutations assigned to DYS389II were carefully checked to discriminate those actually occurring in the DYS389I fragment from those occurred in the DYS389B fragment. Discrimination was always possible except for data from Heyer et al,11 which were excluded for this locus.
Statistical analysis
Population data were analyzed to obtain estimates of relative mutation rates between pairs of loci from allele repeat count variance and homozygosity. The relationship between relative mutation rates and meiosis-based mutation rates was assessed by logistic regression using loci with both population and meiosis data. Inferred relationship was then used to predict mutation rates for all loci, including those lacking of meiosis data. Analysis procedure is detailed below.
First, we selected one locus to serve as reference (ie, mutation rates for all other loci will be relative to this one). As mentioned above, not all individuals are genotyped for the same set of loci (cf. Supplementary Table S1), and thus it is not possible to use the whole data set in the logistic regression (although data from unused loci will be useful for predictions, see below). As a consequence, a reference locus has to be chosen in a way to maximize the amount of information used (ie, to maximize the number of loci with meiosis participating in the regression analysis). In other words, the reference locus has to be the one that shares genotype data with the greatest number of loci with meiosis. To achieve this, we used the following criteria (in this order): (i) there should be meiosis data for the reference locus; (ii) the number of loci with meiosis data (for at least 100 transmissions) and genotyped in individuals (from the population data) also genotyped for the reference locus should be maximum; (iii) the number of loci genotyped in individuals also genotyped for the reference locus should be maximum; and (iv) the sampling size (number of individuals from population data) of the reference locus should be maximum. Note that the choice of the reference locus influences only the amount of data used in the analysis. Otherwise, the reference locus only sets an arbitrary scale to the relative mutation rates calculated from genetic diversity indices.
Relative mutation rate (R=μl/μr) for each locus l was estimated exclusively from individuals genotyped for both locus l and reference locus r. This ensures that the genetic diversity of the sample of both loci has been influenced by the same demographic history (this allows assuming the same effective population sizes). Thus, in the absence of selection, the differences in genetic diversity can be attributed solely to the mutation process. Moreover, because of the complete linkage of Y-STRs, data for both loci will also share the same exact genealogy (even if a selective process was in action). As both loci have the same genealogy, estimates of the mutation rate ratios will be more accurate than in unlinked loci whose genealogies would vary largely due to the randomness of the coalescent process (eg, nuclear STRs compared in Chakraborty et al17). Estimates θ̂V, l, θ̂V, r, θ̂H, l and θ̂H, r were obtained from repeat count variance and homozygosity for loci l and r (using Equations 1 and 2) and two estimates of the relative mutation rate were calculated from ratios R̂V=θ̂V, l/θ̂V, r and R̂H=θ̂H, l/θ̂H, r.
A number of loci (24 out of 110, see Supplementary Table S1), for which there is population data available, were not genotyped at the reference locus in any of the samples. For those loci, relative mutation rates were estimated as described above but using the total number of individuals available for each locus (we will denote these estimates R̂′V and R̂′H). It must be noted that R̂′V and R̂′H might have a larger error than R̂V and R̂H because the effects of demography are more loosely accounted for. For this reason, they were not used for the estimation of the logistic regression model but only in the prediction of mutation rates (see details below).
A generalized linear model (binary logistic regression18) was applied to the proportion of mutations per meiosis. We tested for the relationship between meiosis mutation rate and population relative mutation rates (R̂V and R̂H). Besides, some studies have proposed that microsatellite mutation rates depend on allele length,19, 20 motif size and motif structure.19 Thus, in addition to R̂V and R̂H, mean allele repeat count (A: estimated from the population data), CG content in motif (PCG: proportion of CG base pairs in the motif), and the categorical variables motif size (M: tri-, tetra-, penta- or hexanucleotide motif) and repeat structure (S: simple versus complex) were considered explanatory variables. Information about Y-STR motifs was obtained from Kayser et al, Järve et al, Gusmão et al and Leat et al.21, 22, 23, 24
Problems of multicollinearity were evaluated on the full model (containing all explanatory variables), as collinear variables represent partial redundant information and correlations between variables generate unreliable individual estimates of regression coefficients. Alternative models obtained after removing different combinations of collinear variables were considered and reduced by stepwise removal of variables to minimize Akaike information criterion (AIC, ie, a standard procedure to find the explanatory variable combination, which accounts for the maximum of the variability with the minimum number of variables). Reduced models were hereafter compared through their pseudo-R2 value (calculated by the maximum likelihood method25). Pseudo-R2 measures the amount of variation in the observed mutation rates explained by the model. The reduced model with the highest pseudo-R2 was chosen to predict mutation rates for all loci. As explained before, for loci whose R̂V and R̂H could not be calculated, R̂′V or R̂′H was used as a proxy (estimates for those loci will be distinguished in the results, as they are theoretically less reliable).
All statistical analyses were performed in R,26 using the packages binom27 for calculation of CI, ape28 for calculation of heterozygosity and pscl29 for calculation of pseudo-R2. A script in R language with the detailed analysis is available from the authors upon request.
Validation of the approach
Performance of the statistical approach proposed was evaluated by means of simulations. In each simulation, a set of 108 fully linked loci was considered. Loci were divided in three motif size categories: 36 ‘tri’, 36 ‘penta’ and 36 ‘tetra’. ‘Tri’ loci evolved at six different mutation rates (10−4, 2 × 10−4, 4 × 10−4, 8 × 10−4, 1.6 × 10−3 and 3.2 × 10−3, measured in mutations per generation). ‘Penta’ loci evolved at mutation rates double to those for ‘tri’ loci (ie, 2 × 10−4, 4 × 10−4, 8 × 10−4, 1.6 × 10−3, 3.2 × 10−3 and 6.4 × 10−3) and ‘tetra’ loci evolved at mutation rates quadruple to those for ‘tri’ loci (ie, 4 × 10−4, 8 × 10−4, 1.6 × 10−3, 3.2 × 10−3, 6.4 × 10−3 and 1.28 × 10−2). Note that categories ‘tri’, ‘penta’ and ‘tetra’ are arbitrary (both in their name and their influence in mutation rate) and are only used to include the effect of a categorical variable in the evaluation of the proposed approach. For each mutation rate within each locus category, six loci differing in the amount of observed meiosis (ie, 50, 150, 500, 1500, 5000 and 15000 meiosis) have been considered. To sum up, three categories times six mutation rates, times six loci differing in the number of meiosis gives 108 total simulated loci.
Meiosis data were simulated using the binomial distribution, with the probability equal to the true mutation rate and the number of observations to the number of meiosis. Population data were simulated with the coalescent simulator SimCoal230 under an SMM. A sample size of 500 haplotypes was taken from a single population of constant effective size of 1500 individuals (this effective size combined with the simulated mutation rates yielded genetic diversity levels similar to those found on Y-STRs, ie, around 2–14 alleles per locus).
Mutation rates estimates were obtained for each locus either by using exclusively meiosis data or by using a logistic regression on the observed mutations in meiosis using R̂H and the simulated categorical variable (‘motif size’) as explanatory variables, according to the final model chosen with the real data (see results). The process was repeated 10 000 times. Root of the relative mean squared error ( , where n is the number of simulations, μ̂i is the estimated mutation rate in simulation i and μ is the true mutation rate) was calculated for the two types of mutation rate estimates at each of the 108 loci.
, where n is the number of simulations, μ̂i is the estimated mutation rate in simulation i and μ is the true mutation rate) was calculated for the two types of mutation rate estimates at each of the 108 loci.
Results
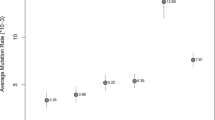
Locus DYS643 was selected as reference locus following the criteria described above. Mutation rates relative to reference locus were estimated from repeat count variance and homozygosity for 86 loci, which were used in the logistic regression model (Supplementary Table 1). Problems of multicollinearity were found between R̂V and the mean repeat count (A), between R̂H and A and between R̂H and CG content in motif (PCG). Thus, we considered three alternative models with a different combinations of noncollinear variables each: model m1 including R̂H plus the motif size (M) and repeat structure (S); model m2 including R̂V plus M, content in motif (PCG) and S; m3 including A plus M, PCG and S. The AIC minimization approach led to the removal of variable PCG in m2 and m3. Final models (Supplementary Tables S3, S4 and S5) were ranked by their pseudo-R2 values: 0.84 for reduced m1, 0.83 for reduced m2 and 0.67 for reduced m3. Reduced m1 model was therefore selected to make predictions on mutation rates from population data for all loci, using R̂H or R̂′H. Reduced m1 (Lμ=β0+β1R̂H+β2Mtri+β3Mtetra+β4Mpenta+β5Ssimple+error; Supplementary Table S3 and Figure 1) shows that mutation rate estimated from meiosis (Lμ) increases with R̂H (ie, β1>0), depends on repeat size (highest for tetranucleotide loci followed by penta- and tri-, ie, β3>β4>β2) and on the complexity of the loci (higher for simple than for complex loci, ie, β5>0). Note that the coefficient of categorical variables is a value relative to the coefficient of the category no explicitly represented in the equation (ie, hexanucleotide repeat motif class and the complex structure class).
Mutation rate estimates (measured in mutations per generation) from meiosis for 80 Y-STR loci (points) and prediction from logistic regression for the eight categories of loci defined by motif size and repeat structure (lines). Continuous lines represent the predictions for loci with a simple repeat structure and dashed lines for complex loci. Thick black lines are used for the predictions of tetranucleotide loci, thick gray lines for hexa- loci, thin black lines for penta- loci and thin gray lines for tri- loci. The logistic regression model (Lμ=−6.863+0.539R̂H−1.176Mtri+0.478Mtetra−1.130Mpenta+0.236Ssimple+error, see Supplementary Table S3 for coefficient P-values) gives the relationship between the logit of mutation rate (Lμ) and the predictive variables R̂H (population relative mutation rate estimated using homozigosity), M (motif size: tri-, tetra-, penta- and hexanucleotide classes) and S (repeat structure: simple or complex). The model shows that Lμ increases with R̂H and depends on repeat size (highest for tetranucleotide loci followed by hexa-, penta- and tri- in this order) and on the complexity of the loci (higher for simple than for complex loci).
For comparison, results from simple models (ie, including each explanatory variable separately) are reported in Supplementary Table S6. They show that all explanatory variables, but the repeat structure, explain significantly part of the variability of mutation rate estimates, although during the model minimization process some were excluded because they provide redundant or nonindependent information. Although repeat structure is not able to significantly explain mutation rate variability when it is the only explanatory variable, it is found to provide significant information when analyzed in combination with other explanatory variables (Supplementary Tables S3, S4 and S5).
Predicted values for mutation rates range from 3.60 × 10−4 mutations per generation for DYS645 to 9.64 × 10−3 for DYS449 (average 2.12 × 10−3, SD=1.58 × 10−3; Supplementary Table 1). For those loci that are not genotyped in any individual genotyped for the reference locus in the population data (see Supplementary Table 1), differences in population history and genealogies are expected to make an additional contribution to the variance in mutation rate estimates, although this does not seem to be too important (exclusion of those loci hardly changes the average predicted mutation rate to 2.25 × 10−3, SD=1.65 × 10−3). In total, regression approach provides an estimate for 71 loci with either zero observed mutations in meiosis (ie, point estimate of mutation rate was zero) or lacking meiosis observations.
It is worth to notice that 45 out of 80 loci with meiosis data share their meiosis mutation rate estimates and CI with at least another locus (given that often the same number of mutations are observed in the same number of meiosis), while mutation rates predicted by regression are different from each other for all loci. Simulations showed that the error associated with meiosis mutation rate estimates is strongly influenced by the number of meiosis, whereas the error of regression estimates seems independent of the number of observed meiosis (Figure 2 reports results for the four simulated mutation rates shared by all loci categories, see Materials and methods). Error in both estimates depends on the true mutation rate, decreasing for higher true mutation rates. However, this decrease is stronger for regression estimates than for meiosis estimates. An interesting feature is that regression estimates are more accurate than meiosis estimates when a low number of meiosis is available, but the contrary occurs for high number of meiosis observations. Although this general pattern seems to be independent of the true mutation rate, the threshold from which meiosis estimates are more accurate than regression estimates increases with the true mutation rate. It is important to remember that the behavior described by simulations concerns only loci for which a meiosis estimate is available; however, the regression approach provides an estimate even when meiosis data are not available.
Root of the relative mean squared error (RrelMSE) for mutation rate estimates calculated from meiosis data (filled circles) or from regression (open circles, triangles and squares) at each of 108 simulated loci. RrelMSE for estimates for loci mutating at true mutation rates of (a) 4 × 10−4 mutations per generation, (b) 8 × 10−4, (c) 1.6 × 10−3 and (d) 3.2 × 10−3. RrelMSE for regression-based estimates also depends on the category the loci belong to: ‘tri’ (triangles), ‘tetra’ (squares) or ‘penta’ (open circles).
Discussion
Mutation rates are expected to vary substantially across Y-microsatellite loci (Zhivotovsky et al31 and references therein). Such large variation has been attributed to motif size, complexity of repeat structure and allele size (eg, Gusmão et al, Kayser et al, Carvalho-Silva et al, Dupuy et al and Ge et al12, 21, 32, 33, 34). Our results are in general agreement with the aforementioned works. We found that meiosis mutation rates are positively correlated with population diversity (estimated by either homozygosity or relative repeat count variance, Supplementary Tables S3 and S4) and mean repeat count (Supplementary Table S5), and depends on repeat motif and repeat structure (Supplementary Table S3). The model selection approach used in this work indicates that a model including relative genetic diversity (from homozygosity), repeat motif and repeat structure as predictive variables is the best one to explain the variability found in meiosis-based estimates of mutation rate. However, it should be noted that the alternative models tested (Supplementary Tables S4 and S5) are valid too, although their lower pseudo-R2 values indicate that they might have a lower performance for making inferences.
Correlation between mutation rates from meiosis and the relative mutation rates based on homozygosity was positive and highly significant (Supplementary Tables S3 and S6). The latter is estimated from population data, and thus corresponds to the ‘evolutionary’ mutation rates (ie, the effective mutation rate integrated over the history or gene tree of the sample). Pedigree-based mutation rate estimates have been shown to be up to 10-fold higher than evolutionary mutation rate estimates (for sequence data), not only in Y chromosomes (eg, Zhivotovsky et al31 and Forster et al35) but also in mitochondrial loci (eg, Macaulay et al36 and Heyer et al37). The reasons for this discrepancy are still under discussion and are likely to be found in the different temporal scale of estimation. In fact, slowly mutating loci or reverse mutations and demographic fluctuations or differential selection over generations are expected to affect population-based diversity (see discussion in Zhivotovsky et al31). It must be noted that our reported estimates (predicted from the logistic model) correspond to the point estimates of mutation rates (ie, mutation occurrence in single generation).
Tri-, tetra- and pentanucleotide classes are well represented in the analyzed locus set (with 17, 55 and 7 loci, respectively), whereas hexanucleotide class did not contribute much to the regression model because it is present with only one locus (DYS448) with meiosis observations. We found that the value for the model coefficient (β) is much lower for tri- and pentanucleotide loci than for tetranucleotide loci (Supplementary Tables S3 and S4), which corresponds to general lower mutation rates for tri- and pentanucleotide loci than for tetranucleotide loci (Supplementary Table 1). Such a different behavior is congruent with the results of previous studies. Järve et al22 recently showed that pentanucleotide markers have two times lower repeat variance and diversity than tetranucleotide markers, a feature probably related to a lower occurrence of replication slippage with longer repeats. Regarding trinucleotide markers, Kayser et al21 found that these had often lower variance than tetranucleotide markers, probably because of the effect of low absolute repeat allele lengths included in their sample.21 Lower mutability of shorter alleles compared with longer ones has been observed several times.32, 33, 12, 34 Accordingly, our results show that the variation in meiosis mutation rates could be significantly explained by mean repeat count (Supplementary Tables S5 and S6). Furthermore, when no diversity variable is included in the model, both repeat count and repeat motif contribute independently to explain the mutation rate variability (ie, model m3, Supplementary Table S5).
The repeat structure explains very little the observed STR mutation rates (Supplementary Table S3), but it is maintained after model reduction using the AIC. However, the coefficient β for simple loci is positive in the final reduced m1 (Supplementary Table S3), whereas it is negative when the repeat structure is used as the only explanatory variable (Supplementary Table S6). Thus, the effect of the repeat structure on mutation rate is of difficult interpretation. Previous studies have failed to find a relationship of simple versus complex repeats with genetic diversity among loci.38, 32, 22 These results might be due to the lack of effect of repeat structure. However, our qualitative classification of loci as ‘simple’ or ‘complex’ could be missing essential information of complex loci (ie, differential length of the homogeneous array or combination of variable and constant repeats21) affecting the mutation rate. More precise definitions of the degree of complexity, similar to those used in Kayser et al,21 could yield different results, but require detailed information on loci not readily available.
The model considered in this work for microsatellite evolution (SMM) predicts single-repeat-unit mutational changes. However, violations of this assumption have been reported both in phylogenetic and meiosis studies (eg, Dupuy et al, Ge et al, Forster et al and Nebel et al33, 34, 38, 39), suggesting that more complex models than SMM would better explain microsatellite variation. The ratio of variances in number of repeats between two loci can be still considered a good estimator of the ratio of mutation rates even in case of multistep mutations, provided that deviation from the SMM is similar for both loci (cf. Equation 2 in Chakraborty et al17). Although the same argument is not strictly valid for the estimate of θ from homozygosity, small deviations from the SMM change very little the expected homozygosity (cf. Supplementary Table 1 from Kimmel and Chakraborty9). Only 14 mutations (3.1% of total) involving multiple repeat units are included in meiosis data; therefore, SMM can be considered a reasonable approximation. In addition, the great congruence in prediction between models m1 (using homozygosity) and m2 (using repeat count variance) suggests that the mutation model violation is not an issue for the analysis (results not shown).
Some important outcomes derive from the approach proposed, emphasizing the positive impact of including population polymorphism data for the improvement of mutation rate estimates and the identification of loci distinctiveness. First, mutation rate estimates were obtained for 30 loci lacking estimates from meiosis observations. Second, locus-specific values of mutation rates can be obtained, whereas meiosis-based estimates give often equal values for several loci. Third, estimates can be obtained also for loci with very low mutation rates, for which a large sample of meiosis data is required to obtain a nonzero mutation rate estimate. Regarding this point, this work provides mutation rate estimates for 41 loci whose mutation rate estimate from meiosis was zero because no mutations had been observed. Finally, regression-based estimates present lower error than estimates from meiosis when only a ‘low’ number of meiosis is observed.
The analysis performed in this work represents a valuable tool for selecting most reliable markers to increase Y-STR set currently applied in forensic and kinship analyses. Also, the choice of adequate mutation rates keeps being an issue of great concern when inferences on human diversity and population history are pursued, as put in evidence in a recent work.6 To account for the different variability of microsatellite loci, these authors use repeat count variance to obtain recalibrated evolutionary mutation rates for groups of loci. Our approximation allows more detailed results by achieving an adjusted mutation rate for each locus separately. The same methodology could be used to estimate population or lineage-specific mutation rates, as different lineages and populations are often characterized by specific allele combinations32, 33, 39 and mutation rate seems to be affected by allele size and structure. Finally, the analysis presented here can be easily automated for a database, allowing the updating of estimates when new population and meiosis data are incorporated from upcoming studies.
References
Hanson EK, Ballantyne J : Comprehensive annotated STR physical map of the human Y chromosome: Forensic implications. Leg Med 2006; 8: 110–120.
Kayser M, Sajantila A : Mutations at Y-STR loci: implications for paternity testing and forensic analysis. Forensic Sci Int 2001; 118: 116–121.
Mulero JJ, Chang CW, Calandro LM et al: Development and validation of the AmpFSTR® Yfiler™ PCR amplification kit: a male specific, single amplification 17 Y-STR multiplex system. J For Sci 2006; 51: 64–75.
Lim S, Xue Y, Parkin E, Tyler-Smith C : Variation of 52 new Y-STR loci in the Y Chromosome Consortium worldwide panel of 76 diverse individuals. Int J Legal Med 2007; 121: 124–127.
Vermeulen M, Wollstein A, van der Gaag K et al: Improving global and regional resolution of male lineage differentiation by simple single-copy Y-chromosomal short tandem repeat polymorphisms. Forensic Sci Int Genet 2009; 3: 205–213.
Shi W, Ayub Q, Vermeulen M et al: A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations. Mol Biol Evol 2010; 27: 385–393. msp243.
Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C : A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet 2002; 71: 466–482.
Xue Y, Zerjal T, Bao W et al: Male demography in East Asia: a north-south contrast in human Population expansion times. Genetics 2006; 172: 2431–2439.
Kimmel M, Chakraborty R : Measures of variation at DNA repeat loci under a general stepwise mutation model. Theor Popul Biol 1996; 50: 345–367.
Xu H, Fu Y : Estimating effective population size or mutation rate with microsatellites. Genetics 2004; 166: 555–563.
Heyer E, Puymirat J, Dieltjes P, Bakker E, de Knijff P : Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet 1997; 6: 799–803.
Gusmão L, Sánchez-Diz P, Calafell F et al: Mutation rates at Y chromosome specific microsatellites. Hum Mutat 2005; 26: 520–528.
Gusmão L, Alves C, Beleza S, Amorim A : Forensic evaluation and population data on the new Y-STRs DYS434, DYS437, DYS438, DYS439 and GATA A10. Int J Legal Med 2002; 116: 139–147.
Mulero JJ, Budowle B, Butler JM, Gusmão L : Nomenclature and allele repeat structure update for the Y-STR locus GATA H4. J For Sci 2006; 51: 694.
Butler JM, Reeder DJ : Short tandem repeat DNA internet database, 2009. Available at: http://www.cstl.nist.gov/strbase/.
Wilson EB : Probable inference, the law of succession, and statistical inference. J Am Stat Assoc 1927; 22: 209–212.
Chakraborty R, Kimmel M, Stivers D, Davison L, Deka R : Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc Natl Acad Scie USA 1997; 94: 1041–1046.
Hosmer DW, Lemeshow S : Applied Logistic Regression. 2nd ed. New York, Chichester: Wiley, 2000.
Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B : Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet 1998; 62: 1408–1415.
Ellegren H : Heterogeneous mutation processes in human microsatellite DNA sequences. Nat Genet 2000; 24: 400–402.
Kayser M, Kittler R, Erler A et al: A comprehensive survey of human Y-chromosomal microsatellites. Am J Hum Genet 2004; 74: 1183–1197.
Järve M, Zhivotovsky LA, Rootsi S et al: Decreased rate of evolution in Y chromosome STR loci of increased size of the repeat unit. PLoS ONE 2009; 4: e7276.
Gusmão L, Butler JM, Carracedo A et al: DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci Int 2006; 157: 187–197.
Leat N, Ehrenreich L, Benjeddou M, Cloete K, Davison S : Properties of novel and widely studied Y-STR loci in three South African populations. Forensic Sci Int 2007; 168: 154–161.
Long JS : Regression Models for Categorical and Limited Dependent Variables. Thousand Oaks, California, USA: Sage Publications Inc, 1997, pp 297.
R Development Core Team: R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2009. Available at: http://www.R-project.org.
Dorai-Raj S : binom: binomial confidence intervals for several parameterizations (R package, version 1.0-5), 2009, Available at: http://cran.r-project.org/web/packages/binom/index.html.
Paradis E, Claude J, Strimmer K : APE: analyses of phylogenetics and evolution in R language. Bioinformatics 2004; 20: 289–290.
Jackman S : pscl: Classes and Methods for R Developed in the Political Science Computational Laboratory, Stanford University. Stanford, California: Department of Political Science, Stanford University, 2008. Available at: http://pscl.stanford.edu/.
Laval G, Excoffier L : SIMCOAL 2.0: a program to simulate genomic diversity over large recombining regions in a subdivided population with a complex history. Bioinformatics 2004; 20: 2485–2487.
Zhivotovsky LA, Underhill PA, Cinnio lu C et al: The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 2004; 74: 50–61.
Carvalho-Silva DR, Santos FR, Hutz MH, Salzano FM, Pena SD : Divergent human Y-chromosome microsatellite evolution rates. J Mol Evol 1999; 49: 204–214.
Dupuy BM, Stenersen M, Egeland T, Olaisen B : Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum Mutat 2004; 23: 117–124.
Ge J, Budowle B, Aranda XG et al: Mutation rates at Y chromosome short tandem repeats in Texas populations. Forensic Sci Int Genet 2009; 3: 179–184.
Forster P, Röhl A, Lünnemann P et al: A short tandem repeat-based phylogeny for the human Y chromosome. Am J Hum Genet 2000; 67: 182–196.
Macaulay VA, Richards MB, Forster P et al: mtDNA mutation rates – no need to panic. Am J Hum Genet 1997; 61: 983–986.
Heyer E, Zietkiewicz E, Rochowski A et al: Phylogenetic and familial estimates of mitochondrial substitution rates: study of control region mutations in deep-rooting pedigrees. Am J Hum Genet 2001; 69: 1113–1126.
Forster P, Kayser M, Meyer E et al: Phylogenetic resolution of complex mutational features at Y-STR DYS390 in aboriginal Australians and Papuans. Mol Biol Evol 1998; 15: 1108–1114.
Nebel A, Filon D, Hohoff C et al: Haplogroup-specific deviation from the stepwise mutation model at the microsatellite loci DYS388 and DYS392. Eur J Hum Genet 2001; 9: 22–26.
Acknowledgements
We thank Frantz Depaulis for his support and suggestions and Joaquín Navascués for its critical review of the paper. Two anonymous reviewers have contributed to improve the paper. This work was developed during a postdoctoral stay of CB (Bourse pour chercheurs étrangers de la Marie de Paris 2007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Burgarella, C., Navascués, M. Mutation rate estimates for 110 Y-chromosome STRs combining population and father–son pair data. Eur J Hum Genet 19, 70–75 (2011). https://doi.org/10.1038/ejhg.2010.154
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.154
Keywords
This article is cited by
-
Bidirectional dispersals during the peopling of the North American Arctic
Scientific Reports (2023)
-
Large-scale pedigree analysis highlights rapidly mutating Y-chromosomal short tandem repeats for differentiating patrilineal relatives and predicting their degrees of consanguinity
Human Genetics (2023)
-
Assessing 23 Y-STR loci mutation rates in Chinese Han father–son pairs from southwestern China
Molecular Biology Reports (2020)
-
Haplotypic polymorphisms and mutation rate estimates of 22 Y-chromosome STRs in the Northern Chinese Han father–son pairs
Scientific Reports (2018)
-
Whole-sequence analysis indicates that the Y chromosome C2*-Star Cluster traces back to ordinary Mongols, rather than Genghis Khan
European Journal of Human Genetics (2018)