Abstract
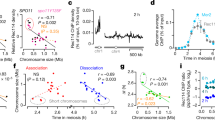
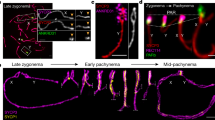
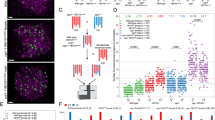
Humans suffer from high rates of fetal aneuploidy, often arising from the absence of meiotic crossover recombination between homologous chromosomes1. Meiotic recombination is initiated by double-strand breaks (DSBs) generated by the SPO11 transesterase2. In yeast and worms, at least one buffering mechanism, crossover homeostasis, maintains crossover numbers despite variation in DSB numbers3,4,5,6,7,8. We show here that mammals exhibit progressive homeostatic control of recombination. In wild-type mouse spermatocytes, focus numbers for early recombination proteins (RAD51, DMC1) were highly variable from cell to cell, whereas foci of the crossover marker MLH1 showed little variability. Furthermore, mice with greater or fewer copies of the Spo11 gene—with correspondingly greater or fewer numbers of early recombination foci—exhibited relatively invariant crossover numbers. Homeostatic control is enforced during at least two stages, after the formation of early recombination intermediates and later while these intermediates mature towards crossovers. Thus, variability within the mammalian meiotic program is robustly managed by homeostatic mechanisms to control crossover formation, probably to suppress aneuploidy. Meiotic recombination exemplifies how order can be progressively implemented in a self-organizing system despite natural cell-to-cell disparities in the underlying biochemical processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hassold, T., Hall, H. & Hunt, P. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 16 (Spec No. 2), R203–R208 (2007).
Keeney, S. in Recombination and Meiosis (eds Egel, R. & Lankenau, D-H.) 81–123 (Springer, 2007).
Chen, S. Y. et al. Global analysis of the meiotic crossover landscape. Dev. Cell 15, 401–415 (2008).
Hillers, K. J. & Villeneuve, A. M. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 13, 1641–1647 (2003).
Martini, E., Diaz, R. L., Hunter, N. & Keeney, S. Crossover homeostasis in yeast meiosis. Cell 126, 285–295 (2006).
Roig, I. & Keeney, S. Probing meiotic recombination decisions. Dev. Cell 15, 331–332 (2008).
Youds, J. L. et al. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327, 1254–1258 (2010).
Rosu, S., Libuda, D. E. & Villeneuve, A. M. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science 334, 1286–1289 (2011).
Jones, G. H. & Franklin, F. C. Meiotic crossing-over: obligation and interference. Cell 126, 246–248 (2006).
Cohen, P. E., Pollack, S. E. & Pollard, J. W. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr. Rev. 27, 398–426 (2006).
Anderson, L. K., Reeves, A., Webb, L. M. & Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151, 1569–1579 (1999).
Holloway, J. K., Booth, J., Edelmann, W., McGowan, C. H. & Cohen, P. E. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4, e1000186 (2008).
Baudat, F., Manova, K., Yuen, J. P., Jasin, M. & Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000).
Kauppi, L. et al. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331, 916–920 (2011).
Mahadevaiah, S. K. et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271–276 (2001).
Larocque, J. R. & Jasin, M. Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol. Cell Biol. 30, 1887–1897 (2010).
Zhang, L., Kleckner, N. E., Storlazzi, A. & Kim, K. P. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc. Natl Acad. Sci. USA 108, 20036–20041 (2011).
Kleckner, N. et al. A mechanical basis for chromosome function. Proc. Natl Acad. Sci. USA 101, 12592–12597 (2004).
Stahl, F. W. & Foss, H. M. A two-pathway analysis of meiotic crossing over and gene conversion in Saccharomyces cerevisiae. Genetics 186, 515–536 (2010).
de Boer, E., Stam, P., Dietrich, A. J., Pastink, A. & Heyting, C. Two levels of interference in mouse meiotic recombination. Proc. Natl Acad. Sci. USA 103, 9607–9612 (2006).
Cole, F., Keeney, S. & Jasin, M. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 24, 1201–1207 (2010).
Mancera, E., Bourgon, R., Brozzi, A., Huber, W. & Steinmetz, L. M. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008).
Dernburg, A. F. et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 (1998).
Barchi, M. et al. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 4, e1000076 (2008).
Lange, J. et al. ATM controls meiotic double-strand-break formation. Nature 479, 237–240 (2011).
Goldfarb, T. & Lichten, M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 8, e1000520 (2010).
Mets, D. G. & Meyer, B. J. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139, 73–86 (2009).
Cole, F., Keeney, S. & Jasin, M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol. Cell 39, 700–710 (2010).
Coop, G., Wen, X., Ober, C., Pritchard, J. K. & Przeworski, M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319, 1395–1398 (2008).
Baker, S. M. et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13, 336–342 (1996).
de Boer, E., Dietrich, A. J., Hoog, C., Stam, P. & Heyting, C. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. J. Cell Sci. 120, 731–736 (2007).
Koehler, K. E., Schrump, S. E., Cherry, J. P., Hassold, T. J. & Hunt, P. A. Near-human aneuploidy levels in female mice with homeologous chromosomes. Curr. Biol. 16, R579–R580 (2006).
Ferguson, K. A., Leung, S., Jiang, D. & Ma, S. Distribution of MLH1 foci and inter-focal distances in spermatocytes of infertile men. Hum. Reprod. 24, 1313–1321 (2009).
Lenzi, M. L. et al. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. Am. J. Hum. Genet. 76, 112–127 (2005).
Bellani, M. A., Boateng, K. A., McLeod, D. & Camerini-Otero, R. D. The expression profile of the major mouse SPO11 isoforms indicates that SPO11β introduces double strand breaks and suggests that SPO11α has an additional role in prophase in both spermatocytes and oocytes. Mol. Cell Biol. 30, 4391–4403 (2010).
Heyting, C. & Dietrich, A. J. Meiotic chromosome preparation and protein labeling. Methods Cell Biol. 35, 177–202 (1991).
Dray, E. et al. Molecular basis for enhancement of the meiotic DMC1 recombinase by RAD51 associated protein 1 (RAD51AP1). Proc. Natl Acad. Sci. USA 108, 3560–3565 (2011).
Roig, I. et al. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet. 6, e1001062 (2010).
Neale, M. J., Pan, J. & Keeney, S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057 (2005).
Cohen, J. Statistical Power Analysis for the Behavioral Sciences 2nd edn (Lawrence Erlbaum Associates, Inc., 1988).
Acknowledgements
We thank present and past members of the Jasin and Keeney laboratories for helpful discussions. F.C. was supported by a Ruth L. Kirschstein NRSA (F32HD51392). S.K. is an Investigator of the Howard Hughes Medical Institute. This work was supported by NIH grant HD040916 (to M.J. and S.K.).
Author information
Authors and Affiliations
Contributions
F.C., S.K. and M.J. designed the experiments. F.C., L.K., J.L., I.R. and R.W. carried out experiments. F.C., S.K. and M.J. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 541 kb)
Rights and permissions
About this article
Cite this article
Cole, F., Kauppi, L., Lange, J. et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 14, 424–430 (2012). https://doi.org/10.1038/ncb2451
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2451
This article is cited by
-
Seeding the meiotic DNA break machinery and initiating recombination on chromosome axes
Nature Communications (2024)
-
Molecular mechanisms and regulation of recombination frequency and distribution in plants
Theoretical and Applied Genetics (2024)
-
Crossover patterning in plants
Plant Reproduction (2023)
-
Meiotic sex chromosome inactivation and the XY body: a phase separation hypothesis
Cellular and Molecular Life Sciences (2022)
-
Two telomeric ends of acrocentric chromosome play distinct roles in homologous chromosome synapsis in the fetal mouse oocyte
Chromosoma (2021)