Abstract
ErbB2, an important membrane-bound receptor tyrosine kinase, was discovered nearly 30 years ago, but a natural ligand has never been found previously. ErbB2 is also an important oncogene and anticancer target, and its overexpression in cancer is associated with poor disease prognosis. Here, we report that human prolidase (PEPD) is a high affinity ligand of ErbB2 and binds as a homodimer to subdomain 3 in the extracellular domain of this receptor. In ErbB2-overexpressing cells, both ErbB2 monomers and activated dimers exist. PEPD bound to ErbB2 monomers relatively slowly but caused ErbB2 dimerization, ErbB2 phosphorylation and downstream signaling. In contrast, PEPD bound rapidly to ErbB2 homodimers and rapidly silenced ErbB2 dimer-Src signaling, a key oncogenic pathway of ErbB2, by disrupting the association of Src with ErbB2. PEPD also caused pronounced ErbB2 depletion, resulting from ErbB2 internalization and degradation. Moreover, PEPD strongly inhibited the DNA synthesis, anchorage-independent growth and invasion and migration of cells that overexpressed ErbB2 but had no effect on cells without ErbB2 overexpression. Cells became sensitized to PEPD upon achieving stable ErbB2 overexpression. Thus, the impact of PEPD on ErbB2 is predominantly inhibitory, and PEPD targets cells addicted to ErbB2. PEPD is also a dipeptidase, but its enzymatic function is not involved in ErbB2 modulation. These findings revise our understanding of ErbB2 and PEPD and may be especially important for combating ErbB2-positive cancers.
Similar content being viewed by others
Main
ErbB2, also known as Her2 or Neu, is a member of the ErbB family of membrane-bound receptor tyrosine kinases, which also includes ErbB1, ErbB3 and ErbB4. They have critical roles in many aspects of development and various diseases. ErbB2 is best known for its involvement in human breast cancer. ERBB2 gene amplification occurs in 20–30% of breast cancer and is a strong predictor of poor disease prognosis.1, 2, 3 ErbB2-targeted therapies, particularly humanized monoclonal antibody trastuzumab,4, 5, 6 have shown considerable clinical efficacy. All ErbB receptors have an extracellular domain (ECD), a transmembrane domain and an intracellular domain. Ligand binding to ECD leads to homo or heterodimerization, followed by activation of the intrinsic protein tyrosine kinase, though ErbB3 is kinase-impaired, and tyrosine phosphorylation in the intracellular domain, which recruits and activates other signaling molecules. To date, while multiple ligands have been identified for ErbB1, ErbB3 and ErbB4,7 no ligand is known for ErbB2, although the receptor itself was discovered nearly 30 years ago.8, 9 However, overexpression of ErbB2 leads to ligand-independent dimerization and activation, and ErbB2 is also a preferred dimerization partner for other ligand-bound ErbBs.10, 11
We recently found that human prolidase, also known as peptidase D (PEPD) among several other names, is a ligand of ErbB1.12 Human PEPD is a homodimeric protein (493 amino acids per subunit) and hydrolyzes dipeptides with proline or hydroxyproline at the carboxy terminus.13 However, the dipeptidase activity of PEPD was not involved in ErbB1 activation.12 Moreover, unlike other ligands of ErbB1 and those of ErbB3 and ErbB4,14 PEPD is devoid of an epidermal growth factor (EGF) motif. This led us to question whether PEPD might interact with ErbB2. Our study to answer this question is described below.
Results
Human PEPD binds to subdomain 3 in the human ErbB2 ECD and causes ErbB2 dimerization
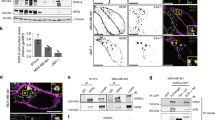
Recombinant human PEPD was generated in bacteria as described recently12 and was incubated at 0.04, 0.2 or 1 μM with 0.04 μM of ErbB2/ECD-Fc (a recombinant chimera of human ErbB2 ECD (Thr23-Thr652) and the Fc fragment of human IgG1) or 0.04 μM of Fc as a control. PEPD bound specifically to the ECD, and each PEPD subunit bound maximally to one copy of ECD (Figure 1a). Given that both PEPD and ErbB2/ECD-Fc (due to Fc) form homodimers in solution, the above result indicates that one PEPD dimer binds to one ECD dimer. PEPD was also incubated at 1 μM with 0.04 μM of ErbB3/ECD-Fc (a chimera of the ECD (Ser20-Thr643) of human ErbB3 and Fc) or 0.04 μM of ErbB4/ECD-Fc (a chimera of the ECD (Gln26-Arg649) of human ErbB4 and the Fc), but no binding could be detected (Figure 1a). As expected, neuregulin-1 (NRG-1) bound to the ECDs of ErbB3 and ErbB4 (Supplementary Figure S1). Thus, PEPD is not a ligand of ErbB3 or ErbB4. We further studied PEPD binding to ErbB2 using Chinese hamster ovary CHO-K1 cells, which expressed a low level of ErbB2 but none of the other ErbBs (Supplementary Figure S2a). Overexpression of human ErbB2 in CHO-K1 cells was readily achieved by gene transfection (Figure 1b). In an enzyme-linked immunosorbent assay (ELISA) using the lysates of ErbB2-overexpressing cells, PEPD bound to ErbB2 with an estimated Kd value of 7.3 nM, whereas there was little PEPD binding to the lysates of control CHO-K1 cells (Figure 1c). PEPD did not bind to the trans-membrane and intracellular regions of ErbB2 (Supplementary Figure S2b). Next, we removed the four ECD subdomains of human ErbB215 one at a time (Figure 1d), using the ErbB2-expressing plasmid pCMV6-XL5-ERBB2 as a template. Similar protein expression levels of ErbB2 and its mutants were detected in CHO-K1 cells transiently transfected with the plasmids (Figure 1d). An equal amount of ErbB2 and its mutants, based on western blotting quantification, were used in the same ELISA mentioned above. Subdomain D1 deletion had little impact on PEPD–ErbB2 binding; the binding affinity after removing subdomains D2 and D4 was reduced 3.5- and 51.0-fold, respectively, but full PEPD binding was achieved by raising the PEPD concentration; removing subdomain D3 completely abolished PEPD binding (Figure 1e). Thus, PEPD bound to D3, but D2 and D4, the latter in particular, might facilitate PEPD binding to ErbB2.
PEPD binds to subdomain 3 of ErbB2 ECD and rapidly promotes ErbB2 dimerization. (a) PEPD at 0.04 μM (+), 0.2 μM (++) or 1 μM (+++) was incubated with ErbB2/ECD-Fc (0.04 μM), ErbB3/ECD-Fc (0.04 μM), ErbB4/ECD-Fc (0.04 μM) or Fc (0.04 μM), pulled down with protein G-sepharose, separated by SDS-PAGE and stained with silver. (b) CHO-K1 cells were transfected with pCMV6-XL5-ERBB2 or the empty vector; 24 h later, cell lysates were prepared and analyzed by western blotting. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used here and elsewhere as a loading control. (c) PEPD binding to ErbB2, as measured by ELISA (n=3), using cell lysates from panel (b); same amount of lysates for all samples (25 μg protein per sample). Error bars indicate S.D. (d) ErbB2 and its mutants; all gene transfections used the same amount of DNA and lasted for 24 h. Cell lysates were analyzed by western blotting. (e) PEPD binding to ErbB2 and its mutants, as measured by ELISA (n=3), using cell lysates from panel (d) and an equal amount of ErbB2 or its mutants. Error bars indicate S.D. (f) CHO-K1 cells stably overexpressing ErbB2 (CHO-K1/ErbB2 cells) were treated with PEPD. PEPD-treated cells and control cells were then treated with BS3 (2 mM, 30 min). Cell lysates were analyzed by western blotting using an ErbB2 antibody
Overexpression of ErbB2 in cell is known to cause spontaneous dimerization. As expected, both monomers and dimers of ErbB2 existed in CHO-K1 cells stably overexpressing ErbB2 (Figure 1f). Cells were treated with cross-linker bis(sulfosuccinimidyl)suberate (BS3) before harvest and western blotting analysis. PEPD apparently underwent two phases of ErbB2 binding: rapid binding of PEPD homodimers to preexisting ErbB2 homodimers (no change in ErbB2 monomer level, decrease in ErbB2 dimer level and formation of heterotetramer of 2 ErbB2s and 2 PEPDs at 10 min of PEPD treatment), followed by binding of PEPD homodimer to ErbB2 monomer, which was apparent at 30 min of PEPD treatment (decrease in ErbB2 monomer level, formation of heterotrimer of 1 ErbB2 and 2 PEPDs, formation of new ErbB2 homodimer, and further increase in heterotetramer of 2 ErbB2s and 2 PEPDs) (Figure 1f). Notably, presence of ErbB2 homodimers not linked to PEPD after PEPD treatment likely resulted from incomplete cross-linking of the two proteins by BS3.
PEPD induces ErbB2 phosphorylation slowly and transiently but causes pronounced and persistent ErbB2 depletion
In CHO-K1/ErbB2 cells which stably overexpressed human ErbB2 and human breast cancer BT-474 cells which constitutively overexpressed ErbB2, two key tyrosine phosphorylation sites on ErbB2, pY1221/1222 and pY1196, were measured. ErbB2 tyrosine phosphorylation at both sites was evidently increased after 0.5–1 h of PEPD treatment at 2.7 nM, peaked at 3 h and largely returned to basal level at 24 h (Figures 2a and b). At higher PEPD concentrations (27 and 270 nM), ErbB2 tyrosine phosphorylation at these sites increased further and lasted longer (Figure 2c). The relatively slow ErbB2 tyrosine phosphorylation induced by PEPD is consistent with the relatively slow PEPD binding to ErbB2 monomer and subsequent dimerization (Figure 1f). PEPD binds to preexisting ErbB2 dimer rapidly (Figure 1f), but it is known that such dimers are auto-tyrosine phosphorylated. As expected, neither EGF nor NRG-1 (ligands of other ErbBs) activated ErbB2 in CHO-K1/ErbB2 cells (Supplementary Figure S3). In cells treated with PEPD at 2.7 nM, ErbB2 protein level began to decrease at 1 h and reached its lowest level at 6 h, which was sustained for at least 72 h (Figure 2a), whereas there was no change in ErbB2 level in vehicle-treated cells (Figure 2b). The impact of PEPD on ErbB2 protein level was dose-dependent, and at 270 nM, PEPD caused almost total ErbB2 elimination (Figure 2c). To understand how PEPD caused ErbB2 depletion, we measured ErbB2 ECD in the culture medium and ErbB2-p95 (minus ECD) in the cell lysates after CHO-K1/ErbB2 cells were treated with PEPD, as ErbB2 can undergo ECD shedding. 4-Aminophenylmercuric acid is known to cause ErbB2 ECD cleavage16 and, as expected, generated the p95 fragment (minus ECD) in the cell lysates and the ECD in the medium (Figure 2d). However, even when cells were treated with PEPD at 270 nM for up to 6 h, no ErbB2 ECD shedding occurred (Figure 2d). Thus, PEPD does not cause ErbB2 ECD cleavage. Also, there was no change in ErbB2 mRNA level in either CHO-K1/ErbB2 cells or BT-474 cells after PEPD treatment for 6 h (Figure 2e), indicating that PEPD-induced ErbB2 depletion was not due to inhibition of ERBB2 gene expression either. Next, both CHO-K1 and CHO-K1/ErbB2 cells were treated with vehicle or PEPD, followed by immunofluorescence staining of ErbB2 and PEPD and detection by confocal microscopy. Cells were treated by PEPD at a high concentration (270 nM) to enhance detection. In CHO-K1 cells, ErbB2 staining was negligible, and there was no PEPD staining (Figure 2f), consistent with very low ErbB2 expression in these cells. In CHO-K1/ErbB2 cells, ErbB2 was strongly stained in the plasma membrane, and after incubation with PEPD for 0.25 h, strong PEPD staining in the plasma membrane was also detected, which co-localized with ErbB2 (Figure 2f), consistent with PEPD binding to ErbB2. However, in CHO-K1/ErbB2 cells treated with PEPD for 2 h, staining intensity of both proteins in the plasma membrane decreased, with concurrent increase of staining in the cytoplasm (Figure 2f), suggesting that ErbB2 and PEPD were internalized. ErbB2 was previously shown to undergo clathrin-independent endocytosis, ubiquitination and degradation.17 Indeed, when cells were treated with PEPD at 2.7 nM for 0.5 h, level of ubiquitinated ErbB2 increased significantly (Figure 2g).
ErbB2 activation and depletion by PEPD. (a–c) Cells were treated with PEPD or vehicle; cell lysates were analyzed by western blotting. (d) Cells were treated with or without PEPD (270 nM), using 4-aminophenylmercuric acid (APMA, 1 mM) as a positive control. Cell lysates and media were analyzed by western blotting. (e) Cells were treated with or without PEPD (2.7 nM, 6 h), from which total RNA was isolated for RT-PCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a control. (f) Cells were treated with or without PEPD (270 nM), followed by immunofluorescence staining of ErbB2 and PEPD and confocal microscopy. Scale bar: 10 μm. (g) Cells were transfected with ubiquitin (pMT107-His-Ub) and 24 h later treated with or without PEPD (2.7 nM, 0.5 h). Cell lysates were incubated with an ErbB2 antibody, pulled down with protein G-agarose and analyzed by western blotting
ErbB2 phosphorylation results from direct PEPD binding, but the dipeptidase function of PEPD is not involved
We next asked whether stimulation of ErbB2 phosphorylation by PEPD results from direct interaction between the two proteins. First, CHO-K1 cells were transfected with a kinase-dead ErbB2 mutant (K753M)18 for 24 h and then treated with PEPD at 270 nM for 3 h, a condition shown to cause maximal phosphorylation of wild-type ErbB2. The ErbB2 mutant was overexpressed in the cells after gene transfection, but PEPD failed to stimulate its phosphorylation, whereas under the same experimental condition, PEPD stimulated the phosphorylation of the wild-type ErbB2 (Figure 3a). Next, CHO-K1 cells were transfected with the ErbB2 mutants lacking an ECD subdomain (Figure 1d) for 24 h and then treated with PEPD at 270 nM for 3 h. PEPD-induced ErbB2 phosphorylation at both pY1221/1222 and pY1196 in both ErbB2/delD1 and ErbB2/delD2 was comparable to that in WT-ErbB2, absent in ErbB2/delD3 and attenuated in ErbB2/delD4 (Figure 3a), which correlated well with PEPD binding to these mutants (Figure 1e). These results suggest that ErbB2 activation by PEPD in cells results entirely from direct binding of PEPD to ErbB2.
PEPD directly activates ErbB2, but its dipeptidase function is not involved. (a) CHO-K1 cells were transfected with ErbB2 or a mutant (all carried by pCMV6-XL5) and 24 h later treated with vehicle or PEPD. Cell lysates were analyzed by western blotting. (b) CHO-K1 cells were transfected with ErbB2 and 24 h later treated with vehicle, PEPD or its mutants. Cell lysates were analyzed by western blotting
Four mutants of human PEPD were generated and evaluated to better understand PEPD as an ErbB2 ligand, including R184X-PEPD (deletion of 309 amino acids from the C-terminus), R265X-PEPD (deletion of 228 amino acids from the C-terminus), G278D-PEPD (G→D at amino acid no.278) and X265R-PEPD (deletion of 228 amino acids from the N-terminus) (Supplementary Figure S4a). CHO-K1 cells were transfected with wild-type human ErbB2 for 24 h and then treated with wild-type PEPD and each of its mutants at 270 nM for 3 h. The PEPD mutants failed to induce ErbB2 phosphorylation, except G278D-PEPD which was identical to WT-PEPD in activating ErbB2 (Figure 3b). However, G278D-PEPD is enzymatically inactive.19 Interestingly, only WT-PEPD and G278D-PEPD could form homodimers (Supplementary Figure S4b). Thus, the dipeptidase activity of PEPD is not involved in ErbB2 activation, but homodimerization of PEPD likely is required for PEPD to bind and activate ErbB2.
Notably, in the experiments described above, PEPD or its G278D mutant did not cause decrease in ErbB2 protein level, whereas PEPD caused pronounced ErbB2 depletion in cells stably or constitutively overexpressing ErbB2 (Figures 2a and c). We found that in CHO-K1 cells transiently overexpressing ErbB2, the majority of ErbB2 molecules resided intracellularly, whereas in CHO-K1 cells stably overexpressing ErbB2, the majority of ErbB2 molecules were expressed on cell surface (Supplementary Figure S5). This explains why PEPD treatment for 3 h did not cause clear ErbB2 protein decrease in cells transiently overexpressing ErbB2.
Intracellular PEPD does not modulate ErbB2
PEPD is mainly a cytosolic protein. Endogenous PEPD levels in CHO-K1/ErbB2 cells and BT-474 cells were relatively low, but PEPD level could be readily elevated in these cells via gene transfection. Modest or strong PEPD overexpression was detected at 24 h after transfection of a plasmid expressing human PEPD, but neither ErbB2 tyrosine phosphorylation nor ErbB2 protein expression changed following PEPD overexpression (Figure 4a). To find out whether cells release PEPD, PEPD-overexpressing cells and control cells were cultured in medium for 24 h, followed by measurement of PEPD level in the cell lysates by western blotting and in the media by ELISA. PEPD concentrations in the media of PEPD-overexpressing cells were 5.9-fold (CHO-K1/ErbB2 cells) and 10.3-fold (BT-474 cells) higher than in the media of the control cells (Figure 4b). Because all cells appeared morphologically normal and healthy, the above result suggests that PEPD may be actively released by the cells. However, due to apparently excessive dilution by the culture medium, extracellular PEPD concentration was too low (<0.3 nM) to impact ErbB2 (Figure 4b).
No effect of intracellular PEPD on ErbB2, but PEPD is released from cell. (a) Cells were transfected with a plasmid expressing human PEPD (pCMV6-XL5-PEPD) or the empty vector for 24 h. Cell lysates were analyzed by western blotting. (b) Cells were transfected with human PEPD or the empty vector; 24 h later, the cells were washed and cultured in fresh medium (∼1 × 106 cells/2 ml medium) for another 24 h, followed by collection of both cells and the medium. PEPD levels in the cell lysates were measured by western blotting, and PEPD concentration in the medium was measured by ELISA (n=3). Error bars indicate S.D.
PEPD rapidly silences ErbB2-Src signaling
ERK lies downstream of ErbB2. As expected, PEPD treatment (2.7 nM) led to ERK activation in a time frame which coincided with that of ErbB2 phosphorylation (compare Figure 5a with Figure 2a), suggesting rapid signal transmission from activated ErbB2 to ERK. Indeed, in CHO-K1 cells expressing the kinase-dead ErbB2 mutant K753M, PEPD caused neither ErbB2 phosphorylation (Figure 3a) nor ERK phosphorylation (Supplementary Figure S6). AKT also lies downstream of ErbB2, but its S473 phosphorylation, which is critical for its function,20 was reduced by PEPD (2.7 nM) in both CHO-K1/ErbB2 cells and BT-474 cells in a time-dependent manner (Figure 5a), whereas vehicle treatment had no impact on AKT phosphorylation (Figure 5b). This suggested that PEPD modulated ErbB2 signaling via additional mechanisms. As mentioned before, ErbB2 overexpression causes spontaneous dimerization and auto-tyrosine phosphorylation. It is also known that Src and PI3K are activated when bound to tyrosine-phosphorylated ErbB2 homodimers or heterodimers, which lead to activation of AKT. In CHO-K1/ErbB2 cells, ErbB2 associated with Src along with CK2, a Src substrate and a pleiotropic serine/threonine protein kinase,21, 22 but PI3K was not detected, whereas in BT-474 cells, ErbB2 associated with Src, CK2 and PI3K (Figure 5c). Treatment of these cells with PEPD (2.7 nM) for only 0.5 h led to marked dissociation of Src and CK2 from ErbB2, but PI3K remained associated with ErbB2 (Figure 5c). Accordingly, Src kinase activity was significantly attenuated by PEPD in both CHO-K1/ErbB2 and BT-474 cells, while PI3K activity remained unchanged (Figure 5d). However, neither Src nor PI3K was significantly modulated by PEPD in CHO-K1 cells (Figure 5d). Further experiments showed that Src phosphorylation at Y419, critical for its kinase function,23 decreased significantly in both CHO-K1/ErbB2 and BT-474 cells after treatment with PEPD (2.7 nM) for only 10 min (Figure 5e), indicating rapid PEPD binding to ErbB2 dimers and rapid disruption of the ErbB2-Src signaling unit. Given that the impact of PEPD on Src occurred before a clear change in ErbB2 phosphorylation and expression, particularly in CHO-K1/ErbB2 cells (Figure 2a), PEPD likely altered the conformation of preexisting ErbB2 dimers, causing Src disassociation from ErbB2.
PEPD silences ErbB2-Src signaling. (a and b) Cells were treated with solvent or PEPD (2.7 nM). Cell lysates were analyzed by western blotting. (c) Cells were treated with or without PEPD (2.7 nM, 0.5 h). Cell lysates were incubated with an ErbB2 antibody, pulled down with protein G-agarose and analyzed by western blotting. (d) Cells were treated with or without PEPD (2.7 nM) and then measured for Src activity and PI3K activity in the cell lysates (n=3). Error bars indicate S.D. (e) Cells were treated with or without PEPD (2.7 nM). Cell lysates were analyzed by western blotting. (f) Cells were treated with pervanadate (30 μM, 10 min) and then treated with or without PEPD (2.7 nM, 0. 5 h). Cell lysates were incubated with a PTEN antibody, pulled down with protein G-agarose and analyzed by western blotting. (g) Cells were treated with solvent or PEPD (2.7 nM, 1 h); cytosolic fraction and membrane fraction were prepared and analyzed by western blotting. Both glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and pro-TGFα (pro-transforming growth factor α) were used as loading controls
PEPD-induced suppression of Src and CK2 was accompanied by loss of PTEN phosphorylation at S380 (Figure 5a), compared with vehicle-treated controls (Figure 5b). PTEN phosphorylation at this site by CK2 contributes to the inhibition of PTEN activity.24 PTEN dephosphorylates membrane-bound PIP3, thereby inhibiting AKT phosphorylation. Src also prevents PTEN translocation to plasma membrane by directly phosphorylating PTEN on tyrosine residues.25 Indeed, PEPD (2.7 nM, 0.5 h) inhibited PTEN tyrosine phosphorylation in both CHO-K1/ErbB2 and BT-474 cells (Figure 5f), and PTEN translocation from cytoplasm to plasma membrane increased after PEPD treatment (2.7 nM, 1 h; Figure 5g).
PEPD targets cells overexpressing ErbB2
Our finding of PEPD binding to ErbB2 and the ensuing changes are summarized in Figure 6a. The rapid inhibition of Src that has a major role in ErbB2 oncogenesis26, 27 along with strong ErbB2 depletion suggested that PEPD might inhibit cells overexpressing ErbB2. The potential inhibitory effects of PEPD on CHO-K1, CHO-K1/ErbB2 and BT-474 cells were evaluated using three assays as described below.
Paradigm of ErbB2 modulation by PEPD, and selective inhibition of cells overexpressing ErbB2 by PEPD. (a) Both monomers and tyrosine-phosphorylated dimers exist in cells overexpressing ErbB2. PEPD binds to ErbB2 as a homodimer; each PEPD subunit binds to one ErbB2 ECD subdomain 3. PEPD rapidly binds to ErbB2 dimers, silencing ErbB2-Src signaling by causing Src disassociation from ErbB2, but PI3K is not affected. PEPD binds to ErbB2 monomers somewhat slowly but causes ErbB2 dimerization and phosphorylation, leading to activation of downstream signaling. PEPD also causes strong and persistent ErbB2 depletion resulting from PEPD-induced ErbB2 internalization and degradation. (b) Cells growing in six-well plates were treated with vehicle or PEPD for 48 h and then measured for DNA synthesis by BrdU incorporation. (c) Cells growing in soft agar in six-well plates were treated with vehicle or PEPD for 21 days (with medium change every 3–4 days) and then examined for colony formation. (d) Cells growing in invasion chambers were treated with vehicle or PEPD for 48 h; cells that invaded through a Matrigel membrane were counted. Error bars in panels (b–d) indicate S.D. (n=3)
The effect of PEPD on DNA synthesis was measured by BrdU incorporation via flow cytometry. Cells were treated by PEPD at 2.7 and 27 nM for 48 h. Although PEPD was ineffective in CHO-K1 cells, it reduced the number of BrdU-positive CHO-K1/ErbB2 cells and BT-474 cells by up to 65 and 50%, respectively (Figure 6b). To measure the effect of PEPD on anchorage-independent growth, cells were grown in soft agar and treated with PEPD at 2.7 and 27 nM for 21 days, with medium change every 3–4 days. Colony formation was not significantly affected by PEPD in CHO-K1 cells but was markedly inhibited by PEPD in both CHO-K1/ErbB2 and BT-474 cells. PEPD reduced the number of colonies with diameter of≥100 μm in CHO-K1/ErbB2 and BT-474 cells by up to 50 and 57%, respectively (Figure 6c). The effect of PEPD on cell invasion and migration was measured using BD BioCoat Matrigel Invasion Chambers. Cells were grown in the upper chamber and treated with solvent or PEPD at 2.7 and 27 nM for 48 h, and the cells that invaded through the Matrigel membrane at the bottom of the upper chamber were counted. PEPD was ineffective in CHO-K1 but inhibited the invasion and migration of CHO-K1/ErbB2 and BT-474 cells by up to 50 and 51%, respectively (Figure 6d). Collectively, these results confirm that the impact of PEPD on ErbB2 is predominantly inhibitory and also show that PEPD selectively targets cells overexpressing ErbB2.
Discussion
Our present discovery breaks the long spell of ErbB2 as an orphan receptor. Compared with other ligands of ErbB receptors, several unusual characteristics of human PEPD are worth highlighting: it does not have an EGF motif; it apparently binds to ErbB2 as a homodimer; and it suppresses the oncogenic signaling of ErbB2. PEPD binds to both ErbB2 monomer and ErbB2 dimer, but as shown in Figure 1f, PEPD binds to ErbB2 homodimers more rapidly than to ErbB2 monomers. This explains why PEPD causes vary rapid disruption of ErbB2-Src signaling (Figure 5) but somewhat slow ErbB2 phosphorylation (Figure 2a). We previously showed that PEPD also binds and activates ErbB1 and that ErbB1 tyrosine phosphorylation induced by PEPD was also slow, peaked after approximately 4 h of PEPD treatment.12 It seems likely that PEPD may also bind slowly to ErbB1 monomer. However, whereas PEPD-induced decrease in ErbB1 protein level was slow (no decrease after 5 h of PEPD treatment, while significantly decreased at 24 h),12 ErbB2 protein level began to decrease after only 1 h of PEPD treatment and reached to its lowest level at 6 h, which was sustained for at least 72 h (Figure 2a). The difference between ErbB1 and ErbB2 with regard to the speed of their depletion by PEPD may be related to the mechanism of their internalization. ErbB1 molecules that are internalized via clathrin-mediated endocytosis are recycled to the cell surface, while clathrin-independent internalization commits the receptor to degradation.28 PEPD-bound ErbB2 likely is internalized via a clathrin-independent mechanism. In fact, PEPD treatment caused rapid increase in ubiquitinated ErbB2 (Figure 2g).
When evaluated for its effect on cell growth and proliferation, PEPD was ineffective in CHO-K1 cells that expressed a low level of ErbB2 but showed strong inhibitory activities in ErbB2-overexpressing CHO-K1 cells (CHO-K1/ErbB2 cells) and BT-474 cells, including inhibition of DNA synthesis, anchorage-independent growth and invasion and migration (Figures 6b–d). This shows that PEPD targets ErbB2 oncogene addiction, as CHO-K1/ErbB2 cells were derived from CHO-K1 cells; at least two mechanisms may be involved: PEPD causes ErbB2 depletion by inducing ErbB2 internalization and degradation, and PEPD inhibits ErbB2-Src signaling by disrupting their association (Figure 5). The effects of PEPD on ErbB2 resemble that of trastuzumab,29, 30 an ErbB2-targeting monoclonal antibody which binds to subdomain 4 of ErbB2 ECD31 and is used currently for treating ErbB2-positive breast cancers. Our preliminary study showed that at equimolar concentrations, inhibition of proliferation of CHO-K1/ErbB2 and BT-474 cells by PEPD or its G278D mutant was similar to, if not better than, that by trastuzumab, while none of the agents showed inhibitory activity in CHO-K1 cells (Supplementary Figure S7). Recombinant human PEPD or its mutant, produced in bacteria, may potentially be a low-cost alternative to the highly expensive trastuzumab, which must be produced in mammalian cells. Further study of the antitumor activity of PEPD is warranted. However, the inhibitory impact of PEPD on cells overexpressing ErbB2 was accompanied by increased ErbB2 phosphorylation and activation of downstream signaling such as ERK (Figures 2a and c and 5a), suggesting that an ErbB2 kinase inhibitor, for example, the dual ErbB1/ErbB2 kinase inhibitor lapatinib,32 may augment the anti-ErbB2 activity of PEPD. Lapatinib may also block ErbB1 activation by PEPD. PEPD does not bind to ErbB3 and ErbB4 (Figure 1a), but it is well known that ErbB2 is a preferred heterodimerization partner with these ErbBs. This raises the question of whether blockade of ErbB2 oncogenic signaling by PEPD may also include inhibition of ErbB2 heterodimer signaling units. Notably, in human breast cancer, the oncogenic activity of ErbB2 depends critically on ErbB3.33, 34
Clearly, we have also discovered a fundamentally new function of human PEPD: an ErbB2 ligand independent of its dipeptidase activity. PEPDs from many species share high sequence homology with human PEPD and may also bind and modulate ErbB2. Although intracellular PEPD does not modulate ErbB2 and PEPD released from cultured cells is too diluted by the medium to impact ErbB2 (Figure 4), PEPD is present in the blood of healthy humans and animals; serum PEPD concentration increased significantly following tissue injury in animals12, 35 and in patients of breast cancer36 and other cancers.37, 38 It is possible that endogenous PEPD released into the extracellular space or blood circulation in vivo may not be unduly diluted, thereby modulating the function of cells and tissues where ErbB2 has an important role physiologically or pathologically.
Materials and Methods
Reagents
Recombinant chimeras, including human ErbB2/ECD-Fc (1129-ER-050), human ErbB3/ECD-Fc (348-RB-050) and human ErbB4/ECD-Fc (1131-ER-050) as well as recombinant Fc of human IgG1 (110-HG-100) were from R&D Systems (Minneapolis, MN, USA). Recombinant human PEPD and its mutants (6xHis tagged to the carboxy terminus) were generated in bacteria and purified using nickel affinity chromatography (Qiagen, Valencia, CA, USA). The purity of each protein was confirmed by gel electrophoresis and silver staining (Supplementary Figure S4b). Details for the preparation of PEPD and its mutants as well as other reagents are provided in the Supplementary Information.
Cell lines and cell culture
BT-474 and CHO-K1 cells were from ATCC (Manassas, VA, USA). CHO-K1/ErbB2 cells were generated by transfecting CHO-K1 cells with pcDNA3-ERBB239 and selected under G418. CHO-K1 and CHO-K1/ErbB2 cells were cultured in F-12K medium (Gibco, Grand Island, NY, USA) supplemented with 10% FBS (Gibco). BT-474 cells were cultured in 50% high-glucose DMEM (Mediatech, Manassas, VA, USA)/50% F-12K medium supplemented with 10% FBS. All cells were cultured in humidified incubators at 37 °C with 5% CO2.
Gene transfection and plasmids
Cells were grown in six-well plates and transfected with a plasmid using FuGENE HD (Promega, Madison, WI, USA) or Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA). pCMV6-XL5-ERBB2 expressing human ErbB2 was generated by cloning full-length human ERBB2 coding sequence to the mammalian expression vector pCMV6-XL5 (Origene, Rockville, MD, USA). All site-directed mutations and deletions in the ERBB2 gene were performed on pCMV6-XL5-ERBB2. Details for plasmid construction are provided in the Supplementary Information. pCMV6-XL5-PEPD expressing wild-type human PEPD was from Origene. pMT107-His-Ub expressing ubiquitin was described previously.40
Western blotting analysis
Preparation of cell lysates, measurement of protein concentration and western blotting analysis were recently described.12 Cell membrane, cytosol fractions or cell lysates minus cell membrane were prepared using the Mem-PER Eukaryotic Membrane Protein Extraction Reagent Kit (Thermo Scientific, Waltham, MA, USA). Cell culture medium was concentrated 20-fold using Centricon (Millipore, Billerica, MA, USA) before analysis. In experiments measuring the binding of PEPD or NRG-1 to ErbB2 ECD, ErbB3 ECD or ErbB4 ECD, a silver staining kit (Invitrogen) was used to display the proteins after gel electrophoresis. To detect ErbB2 receptor dimerization, PEPD-treated cells and control cells were washed with ice-cold PBS and incubated with cross linker BS3 (Pierce, Rockford, IL, USA) at 2 mM for 30 min at room temperature. The cross-linking reaction was terminated by adding 50 mM Tris (final, pH7.5), followed by incubation at room temperature for 15 min. Cell lysates were analyzed by western blotting (3.5% SDS-PAGE).
Immunoprecipitation
PEPD was incubated with ErbB2/ECD-Fc, ErbB3/ECD-Fc, ErbB4/ECD-Fc or Fc in binding buffer for 2 h at 37 °C, followed by pull down with protein G-sepharose beads. The immunoprecipitates were washed with IP washing buffer and analyzed by western blotting. In experiments using whole-cell lysates, cells were lysed in M-PER buffer supplemented with a proteinase inhibitor mix (Roche Applied Science, Indianapolis, IN, USA), and the lysates were incubated with a specific antibody overnight at 4 °C, followed by pull down with protein G-agarose. The immunoprecipitates were washed with IP washing buffer and analyzed by western blotting. To measure the effect of PEPD on PTEN tyrosine phosphorylation, cells were pretreated with 30 μM pervanadate for 10 min to inhibit relevant tyrosine phosphatase25, 30 and then treated with PEPD or vehicle, followed by preparation of cell lysates for analysis. More detail about immmunoprecipitation and preparation of pervanadate is provided in the Supplementary Information.
ELISA-based measurement of PEPD and PEPD binding to ErbB2
Measurement of PEPD by ELISA was recently described.12 To measure PEPD binding to ErbB2 or its mutants, ELISA plates were coated with an ErbB2 antibody (binding to the cytoplasmic tail of ErbB2), and the residual protein binding sites were blocked by 1% BSA in PBS. Serially diluted PEPD was then added to each well, followed by addition of cell lysates (CHO-K1 cells transfected with wild-type human ErbB2 or the empty vector) containing 25 μg of total protein per sample, which was incubated at 37 °C for 2 h. After a wash with PBST, each well was incubated with a biotin-conjugated 6xHis antibody for 2 h at room temperature (note that PEPD is His-tagged). After another round of wash with PBST, each well was incubated with a streptavidin-conjugated horseradish peroxidase (HRP) at room temperature for 45 min. Each well was washed again with PBST, and the bound HRP was measured in a standard colorimetric assay, using 3,3′,5,5′-tetramethylbenzedine as a substrate. In experiments comparing PEPD binding to the wild-type human ErbB2 and its mutants, the lysates of cells transfected with each plasmid were first subjected to western blotting analysis, followed by densitometry measurement of the specific protein bands normalized to a loading control, in order to calculate the amount of lysates that deliver an equal amount of WT-ErbB2 or its mutant (25 μg of total protein/sample was used for the lysates carrying WT-ErbB2). More detail about the ELISA is provided in the Supplementary Information.
Immunofluorescence staining and confocal microscopy
Cells were grown in chamber slides (1.5 × 104 cells/well) overnight, followed by treatment with PEPD or vehicle. The cells were then washed with ice-cold PBS, fixed with 4% paraformaldehyde for 15 min at room temperature, washed again with ice-cold PBS and blocked with 1% BSA in PBS for 45 min at room temperature. The cells were then incubated with an ErbB2 antibody for 1 h at room temperature, washed with PBS, incubated with a FITC-conjugated His-tag antibody (for PEPD detection) and a TRITC-conjugated secondary antibody (for ErbB2 detection) for 1 h at room temperature and washed again with PBS. The cells were then examined with a Zeiss LSM 510 confocal microscope (Thornwood, NY, USA).
RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen), treated with TURBO DNase to remove potential genomic DNA contamination and reverse transcribed into cDNA using the TaqMan Reverse Transcription Reagents (Invitrogen). PCR amplification was carried out with GoTaq Master Mix (Promega) and specific forward and reverse primers. The PCR products were analyzed by electrophoresis with 1% agarose gel, stained by ethidium bromide and visualized under UV light. Further detail is provided in the Supplementary Information.
BrdU assay, soft agar colony-formation assay and cell invasion and migration assay
To measure BrdU incorporation into DNA, cells were grown in six-well plates and treated with PEPD or vehicle for 48 h and then analyzed using the FITC BrdU Flow Kit (BD Pharmingen, San Diego, CA, USA). To measure anchorage-independent colony formation, cells were grown in soft agar in six-well plates and treated with PEPD and vehicle for 21 days, with medium change every 3–4 days. Cell invasion and migration were measured using the BD BioCoat Matrigel Invasion Chambers (BD Biosciences, San Jose, CA, USA). Cells were placed in the upper chambers and treated with PEPD and vehicle for 48 h. Cells that invaded through the Matrigel matrix layer coated on the filter insert that was placed at the bottom of the upper chamber were counted. Further detail for these assays is provided in the Supplementary Information.
PI3 kinase assay and Src kinase assay
PI3K activity and Src kinase activity were measured using the PI3-Kinase Activity ELISA Kit (Echelon, Salt Lake City, UT, USA) and the Universal Tyrosine Kinase Assay Kit (TaKaRa, Mountain View, CA, USA), respectively. PI3K and Src were pulled down from cell lysates with specific antibodies before analysis. Further detail for these assays is provided in the Supplementary Information.
Statistical analysis
Student’s t-test and ANOVA were used for two-group comparison and multi-group comparison, respectively. All tests were two-sided and performed at a nominal significance level of 0.05, that is, P-value of ≤0.05 was considered statistically significant.
Abbreviations
- BS3:
-
bis(sulfosuccinimidyl)suberate
- ECD:
-
extracellular domain
- EGF:
-
epidermal growth factor
- ELISA:
-
enzyme-linked immunosorbent assay
- HRP:
-
horseradish peroxidase
- NRG-1:
-
neuregulin-1
- PEPD:
-
prolidase
References
King CR, Kraus MH, Aaronson SA . Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science 1985; 229: 974–976.
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL . Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177–182.
Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN . The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist 2009; 14: 320–368.
Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002; 20: 719–726.
Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE Jr, Davidson NE et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 2005; 353: 1673–1684.
Dawood S, Broglio K, Buzdar AU, Hortobagyi GN, Giordano SH . Prognosis of women with metastatic breast cancer by HER2 status and trastuzumab treatment: an institutional-based review. J Clin Oncol 2010; 28: 92–98.
Hynes NE, Lane HA . ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5: 341–354.
Schechter AL, Hung MC, Vaidyanathan L, Weinberg RA, Yang-Feng TL, Francke U et al. The neu gene: an erbB-homologous gene distinct from and unlinked to the gene encoding the EGF receptor. Science 1985; 229: 976–978.
Coussens L, Yang-Feng TL, Liao YC, Chen E, Gray A, McGrath J et al. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science 1985; 230: 1132–1139.
Brennan PJ, Kumagai T, Berezov A, Murali R, Greene MI . HER2/neu: mechanisms of dimerization/oligomerization. Oncogene 2000; 19: 6093–6101.
Graus-Porta D, Beerli RR, Daly JM, Hynes NE . ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J 1997; 16: 1647–1655.
Yang L, Li Y, Ding Y, Choi KS, Kazim AL, Zhang Y . Prolidase directly binds and activates epidermal growth factor receptor and stimulates downstream signaling. J Biol Chem 2013; 288: 2365–2375.
Besio R, Alleva S, Forlino A, Lupi A, Meneghini C, Minicozzi V et al. Identifying the structure of the active sites of human recombinant prolidase. Eur Biophys J 2010; 39: 935–945.
Jones JT, Akita RW, Sliwkowski MX . Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett 1999; 447: 227–231.
Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX . Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 2004; 5: 317–328.
Molina MA, Codony-Servat J, Albanell J, Rojo F, Arribas J, Baselga J . Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res 2001; 61: 4744–4749.
Barr DJ, Ostermeyer-Fay AG, Matundan RA, Brown DA . Clathrin-independent endocytosis of ErbB2 in geldanamycin-treated human breast cancer cells. J Cell Sci 2008; 121: 3155–3166.
Fan YX, Wong L, Ding J, Spiridonov NA, Johnson RC, Johnson GR . Mutational activation of ErbB2 reveals a new protein kinase autoinhibition mechanism. J Biol Chem 2008; 283: 1588–1596.
Ledoux P, Scriver CR, Hechtman P . Expression and molecular analysis of mutations in prolidase deficiency. Am J Hum Genet 1996; 59: 1035–1039.
Gao T, Furnari F, Newton AC . PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell 2005; 18: 13–24.
Donella-Deana A, Cesaro L, Sarno S, Ruzzene M, Brunati AM, Marin O et al. Tyrosine phosphorylation of protein kinase CK2 by Src-related tyrosine kinases correlates with increased catalytic activity. Biochem J 2003; 372: 841–849.
Liang K, Esteva FJ, Albarracin C, Stemke-Hale K, Lu Y, Bianchini G et al. Recombinant human erythropoietin antagonizes trastuzumab treatment of breast cancer cells via Jak2-mediated Src activation and PTEN inactivation. Cancer Cell 2010; 18: 423–435.
Bjorge JD, Jakymiw A, Fujita DJ . Selected glimpses into the activation and function of Src kinase. Oncogene 2000; 19: 5620–5635.
Miller SJ, Lou DY, Seldin DC, Lane WS, Neel BG . Direct identification of PTEN phosphorylation sites. FEBS Lett 2002; 528: 145–153.
Lu Y, Yu Q, Liu JH, Zhang J, Wang H, Koul D et al. Src family protein-tyrosine kinases alter the function of PTEN to regulate phosphatidylinositol 3-kinase/AKT cascades. J Biol Chem 2003; 278: 40057–40066.
Muthuswamy SK, Siegel PM, Dankort DL, Webster MA, Muller WJ . Mammary tumors expressing the neu proto-oncogene possess elevated c-Src tyrosine kinase activity. Mol Cell Biol 1994; 14: 735–743.
Sheffield LG . C-Src activation by ErbB2 leads to attachment-independent growth of human breast epithelial cells. Biochem Biophys Res Commun 1998; 250: 27–31.
Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP . Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell 2008; 15: 209–219.
Crowder RJ, Lombardi DP, Ellis MJ . Successful targeting of ErbB2 receptors-is PTEN the key? Cancer Cell 2004; 6: 103–104.
Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 2004; 6: 117–127.
Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW Jr et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 2003; 421: 756–760.
Rusnak DW, Lackey K, Affleck K, Wood ER, Alligood KJ, Rhodes N et al. The effects of the novel, reversible epidermal growth factor receptor/ErbB-2 tyrosine kinase inhibitor, GW2016, on the growth of human normal and tumor-derived cell lines in vitro and in vivo. Mol Cancer Ther 2001; 1: 85–94.
Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF 3rd, Hynes NE . The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA 2003; 100: 8933–8938.
Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res 2008; 68: 5878–5887.
Caliskan A, Yavuz C, Karahan O, Yazici S, Guclu O, Demirtas S et al. Factor-Xa inhibitors protect against systemic oxidant damage induced by peripheral-ischemia reperfusion. J Thromb Thrombolysis 2014; 37: 464–468.
Kir ZO, Oner P, Iyidogan YO, Turkmen S, Kocak H, Koser M et al. Serum prolidase I activity and some bone metabolic markers in patients with breast cancer: in relation to menopausal status. Clin Biochem 2003; 36: 289–294.
Gecit I, Aslan M, Gunes M, Pirincci N, Esen R, Demir H et al. Serum prolidase activity, oxidative stress, and nitric oxide levels in patients with bladder cancer. J Cancer Res Clin Oncol 2012; 138: 739–743.
Arioz DT, Camuzcuoglu H, Toy H, Kurt S, Celik H, Aksoy N . Serum prolidase activity and oxidative status in patients with stage I endometrial cancer. Int J Gynecol Cancer 2009; 19: 1244–1247.
Ding Y, Liu Z, Desai S, Zhao Y, Liu H, Pannell LK et al. Receptor tyrosine kinase ErbB2 translocates into mitochondria and regulates cellular metabolism. Nature Commun 2012; 3: 1271.
Li Y, Paonessa JD, Zhang Y . Mechanism of chemical activation of Nrf2. PLoS One 2012; 7: e35122.
Acknowledgements
We thank Ming Tan of University of South Alabama for providing pCDNA3-ERBB2, Gokul M Das of Roswell Park Cancer Institute for help with BT-474 cells, and Kathy Galus and Michael J Spino Jr of Roswell Park Cancer Institute for assistance with obtaining trastuzumab. This work was supported in part by grants from NIH (R01CA120533, R01CA124627 and R01CA164574).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Raschellà
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Yang, L., Li, Y. & Zhang, Y. Identification of prolidase as a high affinity ligand of the ErbB2 receptor and its regulation of ErbB2 signaling and cell growth. Cell Death Dis 5, e1211 (2014). https://doi.org/10.1038/cddis.2014.187
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.187
Keywords
This article is cited by
-
Depleting receptor tyrosine kinases EGFR and HER2 overcomes resistance to EGFR inhibitors in colorectal cancer
Journal of Experimental & Clinical Cancer Research (2022)
-
Loss of peptidase D binding restores the tumor suppressor functions of oncogenic p53 mutants
Communications Biology (2021)
-
PEPD is a pivotal regulator of p53 tumor suppressor
Nature Communications (2017)
-
The HER2 Signaling Network in Breast Cancer—Like a Spider in its Web
Journal of Mammary Gland Biology and Neoplasia (2014)