Abstract
Necroptosis is a form of programmed cell death that depends on the activation of receptor interacting protein kinase-1 (RIPK1) and RIPK3 by receptors such as tumor necrosis factor (TNF) receptor-1. Structural studies indicate that activation of RIPK3 by RIPK1 involves the formation of oligomers via interactions of the RIP homotypic interaction motif (RHIM) domains shared by both proteins; however, the molecular mechanisms by which this occurs are not fully understood. To gain insight into this process, we constructed versions of RIPK3 that could be induced to dimerize or oligomerize in response to a synthetic drug. Using this system, we find that although the formation of RIPK3 dimers is itself insufficient to trigger cell death, this dimerization seeds a RHIM-dependent complex, the propagation and stability of which is controlled by caspase-8 and RIPK1. Consistent with this idea, we find that chemically enforced oligomerization of RIPK3 is sufficient to induce necroptosis, independent of the presence of the RHIM domain, TNF stimulation or RIPK1 activity. Further, although RIPK1 contributes to TNF-mediated RIPK3 activation, we find that RIPK1 intrinsically suppresses spontaneous RIPK3 activation in the cytosol by controlling RIPK3 oligomerization. Cells lacking RIPK1 undergo increased spontaneous RIPK3-dependent death on accumulation of the RIPK3 protein, while cells containing a chemically inhibited or catalytically inactive form of RIPK1 are protected from this form of death. Together, these data indicate that RIPK1 can activate RIPK3 in response to receptor signaling, but also acts as a negative regulator of spontaneous RIPK3 activation in the cytosol.
Similar content being viewed by others
Main
Necroptosis is a form of programmed cell death that is both mechanistically and morphologically distinct from apoptosis.1, 2 Although apoptosis is defined by the activation of the caspase proteases, necroptosis is triggered by receptor-interacting protein kinase 1 (RIPK1 (Degterev et al.3 and Lin et al.4) and RIPK3.5, 6, 7 Morphologically, necroptosis resembles the unprogrammed process of necrosis, involving cellular swelling and rupture.8 This morphology is distinct from apoptosis, in which dying cells shrink and their contents remain contained within membrane-bound bodies or vesicles. Necroptotic cell death thereby releases cellular contents that are contained during apoptosis; necroptosis is therefore thought to be an inflammatory form of cell death. Consistent with a proposed role in inflammation and immune responses, necroptosis can be triggered by tumor necrosis factor (TNF),2 interferon9 or Toll-like receptor10 signaling, as well as by viral infection via the DNA sensor DAI (DNA-dependent activator of interferon regulatory factor).11 Necroptotic cell death has a role in the host response to viral and bacterial infection,5, 11, 12, 13 as well as the pathogenesis of TNF-induced sterile septic shock.14
The mechanism by which the necroptotic program is initiated has been studied principally in the context of TNF receptor-1 (TNFR1) activation, and it remains incompletely understood. Briefly, ligation of TNFR1 by TNF induces the assembly of a large receptor-proximal complex that includes RIPK1.15 Ubiquitination and phosphorylation events within this complex lead to activation of an nuclear factor-κB transcriptional program and/or MAP kinase activation.15 Subsequently, RIPK1 is deubiquitinated and translocates into the cytosol,16 where it forms additional complexes that have been termed ‘necrosomes’17 or ‘ripoptosomes’;18, 19 these scaffolds support RIPK3 activation, which in turn leads to phosphorylation of the downstream mediator mixed-lineage kinase-like (MLKL)20, 21, 22, 23 and the process of necroptosis. Importantly, the cIAP ubiquitin ligases18, 24 and the pro-apoptotic enzyme caspase-8,25, 26 in concert with its paralog cFLIPL (cellular flice-like inhibitory protein, long isoform), can also be recruited to necrosome complexes, and they antagonize RIPK3 activation and necroptosis.
The assembly and regulation of the RIPK1–RIPK3 necrosome is an open subject of investigation in the field. Recent structural analysis showed that the RIP homotypic interaction motif (RHIM) domains of RIPK1 and RIPK3 form amyloid-like oligomers during RIPK3 activation;17 however, it remains unclear whether RIPK3 oligomerization, RHIM amyloid formation, or both are necessary and/or sufficient for RIPK3 activation. Furthermore, it is unclear how suppressors of necroptosis, such as caspase-8, interact with and regulate RIPK3 oligomers to determine cell fate.
Inducible protein interaction systems have provided fundamental insight into many cellular processes, including cell death. For example, we and others have used versions of the FK506-binding protein (FKBP)–rapamycin interaction system27 to create caspase proteases that could be induced to undergo homo- or heterodimerization by addition of specific drug ligands.28, 29, 30, 31 Here we applied similar strategies to the study of RIPK3, with the goal of defining how its activation is regulated during cell life and in response to stress events that culminate in the induction of necroptosis. Using these systems, we found that dimerization of RIPK3 is able to seed a RHIM-dependent oligomer, the propagation of which is required for induction of necroptosis. This RHIM-dependent oligomerization is directly regulated by RIPK1 and caspase-8. Unexpectedly, we found that although chemical inhibition of RIPK1 inhibited RHIM-dependent RIPK3 oligomerization and cell death, depletion of RIPK1 protein in this system had the opposite effect. Together, these data indicate that RIPK3 oligomerization is both necessary and sufficient for the induction of necroptosis, and that RHIM-dependent oligomerization of RIPK3 recruits capase-8 and RIPK1, which control this process. Further, although RIPK1 is required for receptor-induced activation of RIPK3, we show that it also exerts intrinsic suppression of RIPK3 oligomerization in the cytosol.
Results
RIPK3 dimerization triggers necroptosis, which requires the RHIM domain of RIPK3
In an effort to gain insight into the mechanism by which RIPK3 is activated, we created a chimeric protein composed of murine RIPK3 fused to a single copy of FKBPF36V (hereafter ‘FKBPF36V point mutant (FV) domain’), a protein domain that binds with high affinity to a synthetic bivalent homologue of rapamycin, here called ‘AP1’ (homodimerization drug; Figure 1a). FV domains rapidly dimerize in response to AP1 treatment,27 and we took advantage of this property to investigate the protein–protein interactions involved in RIPK3 activation and necroptosis. Importantly, we chose to append the FV domain to the C-terminus of RIPK3, the same position at which the RHIM domain is located, in an effort to faithfully mimic RHIM-dependent interactions that define RIPK3 activation.17
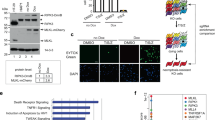
RIPK3 dimerization seeds a RHIM-dependent necrosome complex. (a) Schematic representation of the dimerizable and oligomerizable RIPK3 constructs used in this study. These constructs were cloned upstream of a T2A-GFP sequence, such that RIPK3 constructs contain both N-terminal FLAG and C-terminal 2A epitope tags. (b and c) NIH-3T3 cells stably expressing RIPK3-1xFV were treated with indicated concentrations of AP1 (b), or with 30 nM AP1 in the presence or absence of 200 ng/ml TNFR1-Fc (c), and cell death was assessed over time using an IncuCyte imaging system. (d) NIH-3T3 cells stably expressing RIPK3ΔRHIM-1xFV were treated with increasing doses of dimerizer. (e) NIH-3T3 cells stably expressing RIPK3-1xFV were transfected with indicated siRNAs. Seventy-two hours later cells were treated with 30 nM AP1 and cell death was assessed. *P=0.0001 (f) NIH-3T3 cells stably expressing indicated constructs were treated as indicated, lysed and necrosome complexes were covalently cross-linked using DSS. Resulting complexes were resolved by western blotting. Nec1 and zVAD were used at 30 and 50 μM, respectively, throughout
We expressed RIPK3-1xFV (Figure 1a) in NIH-3T3 cells, a cell line that lacks endogenous RIPK3 expression and is therefore unresponsive to TNF-induced necroptosis (Supplementary Figures S1A and B). We then quantified the cell death responses of NIH-3T3 cells expressing our constructs over time using the IncuCyte imaging system, which allows precise quantification of cell death with high temporal resolution (Supplementary Movies 1 and 2). Expression of RIPK3-1xFV sensitized these cells to necroptosis induced by the combination of TNF and the caspase inhibitor Z-val-ala-asp-(O-methlylated)-fluoromethylketone (zVAD) in a manner analogous to that observed in Jax cells, a murine fibroblast line that expresses endogenous RIPK3 (Supplementary Figure S1C compared with Supplementary Figure S1A). These data indicate that our construct could be faithfully activated by the well-described pathway of TNF receptor-driven cell death. Next, we added AP1 to these cells in the absence of TNF, to test the effect of chemically enforced RIPK3 dimerization in the cytosol. We found that on AP1 addition, these cells underwent limited RIPK3 activation and necroptosis (Figure 1b) in a manner that depended on the concentration of AP1 added. To ensure that the cell death observed on dimerizer addition was not influenced by autocrine TNF production in our cells, we treated them with the TNF blocking reagent TNFR1–immunoglobulin Fc fusion protein (TNFR-Fc). Although TNFR-Fc efficiently inhibited TNF-induced necroptosis in these cells (Supplementary Figure S1D), it did not affect AP1-induced cell death (Figure 1c). However, dimerizer-induced cell death did require the kinase activity of RIPK3 (Supplementary Figure S1E), as well as the downstream mediator of necroptosis MLKL (Figure 1e and Supplementary Figure S2A). Mutation of key phosphorylation sites required for interaction between RIPK3 and MLKL20 also rendered RIPK3-1xFV unable to induce cell death on either TNF or AP1 treatment (Supplementary Figures S1F and I). Together, these data confirm that RIPK3 dimerization leads to necroptosis via direct activation of RIPK3.
The RIP kinases interact via RHIM domains, and mutation of this domain in RIPK3 renders it unresponsive to receptor-driven necroptosis (Supplementary Figure S1G and Li et al.17). We next tested the requirement for this domain in RIPK3 activation via dimerization. To our surprise, given that we were inducing RIPK3 interaction via AP1-mediated homodimerization, we found that versions RIPK3-1xFV in which the RHIM domain was deleted (RIPK3ΔC-1xFV), or in which the key RHIM amino acid sequence VQIG was modified to AAAA (RIPK3ΔRHIM-1xFV) failed to trigger cell death following AP1 treatment (Figure 1d and Supplementary Figures S1G–I). Recent structural evidence demonstrated that the RHIM domain of RIPK3 forms amyloid-like oligomers during RIPK3 activation.17 We therefore hypothesized that although RIPK3 dimerization itself is insufficient for its activation, it may ‘seed’ RHIM oligomers that recruit both RIPK1 and additional molecules of RIPK3, and whose propagation allows RIPK3 activation. To directly test this idea and capture evidence of RIPK3 dimers or oligomers, we performed DSS crosslinking experiments on cells expressing RIPK3ΔRHIM-1xFV or RIPK3-1xFV following dimerizer treatment. Consistent with our hypothesis, we found RIPK3ΔRHIM-1xFV predominantly in a gel-shifted complex that is consistent with dimer formation, whereas RIPK3-1xFV was present in a combination of dimers and larger oligomeric complexes (Figure 1e). We therefore sought to use our constructs to define the regulation of RIPK3 oligomerization and activation.
RHIM-dependent formation of RIPK3 complexes is controlled by RIPK1 and caspase-8
Current models of receptor-driven necroptosis involve the scaffolding and activation of RIPK1 at a plasma membrane receptor, followed by its translocation to the cytosol and the recruitment of both caspase-8 and RIPK3 into a ‘necrosome’ complex;1 RIPK1 can activate RIPK3 in this complex,5 while caspase-8 can suppress this activation.25, 26 We tested whether similar dynamics might influence the receptor-independent formation and propagation of RIPK3 complexes triggered by RIPK3 dimerization. To our surprise, we found that addition of the caspase inhibitor zVAD (Figure 2a), or siRNA-mediated knockdown of caspase-8 (Supplementary Figures S2A and B) notably increased the rate and magnitude of necroptosis triggered by RIPK3-1xFV dimerization, in a manner analogous to that observed with TNF-driven RIPK3 activation in these cells (Supplementary Figure S1B). However, the effects we observed were independent of TNF receptor engagement, as the increased death observed on treatment with AP1 and zVAD were unaffected by the TNF-blocking reagent TNFR1-Fc (Supplementary Figure S2B). Conversely, treatment of cells expressing RIPK3-1xFV with the RIPK1 inhibitor necrostatin-1 (Nec1),3 which blocks TNF-induced necroptosis in these cells (S1C), suppressed RIPK3 dimerization-induced cell death (Figure 2c). These findings indicate that caspase-8 and RIPK1 may intrinsically regulate the initiation and propagation of RIPK3 oligomers in the cytosol, independent of TNF receptor-mediated signaling pathways.
Receptor-independent RIPK3 oligomerization is regulated by RIPK1 and caspase-8. (a–c) NIH-3T3 cells stably expressing RIPK1-1xFV were treated with 30 nM AP1 and the indicated inhibitors, and cell death was assayed by IncuCyte. In b, cells were transfected with indicated siRNAs, then treated with AP1 72 h later. a: *P<0.0001, b: *P=0.0157, c: *P=0.0006. (d) NIH-3T3 cells stably expressing RIPK3-1xFV were treated with 30 nM AP1, as well as Nec1 or zVAD as indicated, and resulting complexes were resolved by western blotting. (e) NIH-3T3 cells expressing the indicated constructs were treated as shown for 30 min, then lysed and subjected to immunoprecipitation using an antibody to the FLAG epitopes expressed on the RIPK3 constructs. Immune complexes were purified and resolved using TruBlot reagents to avoid aspecific signals from immunoglobulins, as described. Nec1 and zVAD were used at 30 and 50 μM, respectively, throughout
To more directly test this idea, we performed DSS crosslinking experiments to visualize the formation and stability of RIPK3 complexes following RIPK3 dimerization, when either caspase-8 or RIPK1 were inhibited (Figure 2d and Supplementary Figure S2C). Consistent with our hypothesis, we found that dimerization of RIPK3 in the presence of zVAD led to an increased shift of RIPK3 into higher-molecular weight complexes consistent with RIPK3 oligomers, while RIPK3 dimerization in the presence of Nec1 diminished the appearance of these complexes. As a further test of these ideas, we immunoprecipitated RIPK3-1xFV or RIPK3ΔRHIM-1xFV from cells treated with AP1 in the presence of Nec1 or zVAD. Previous work has shown that caspase inhibition can stabilize a caspase-8- and RIPK1-containing necrosome complex following TNF treatment,25, 32 and we observed a similar phenomenon following RIPK3 dimerization (Figure 2e and Supplementary Figure S2E). Dimerization itself led to limited recruitment of RIPK1 and caspase-8 to the RIPK3 complex, while addition of zVAD increased association of these proteins. Inhibition of RIPK1 by Nec1, by contrast, eliminated the formation of stable RIPK1- and caspase-8-containing complexes. Similarly, mutation of the RHIM domain of RIPK3 prevented necrosome formation on RIPK3 dimerization. Notably, however, probing crosslinked RIPK3 oligomers with antibodies for RIPK1, caspase-8, or MLKL did not reveal significant association of these proteins with high-molecular weight RIPK3 complexes (Supplementary Figure S2D), consistent with a model in which RIPK3 is the primary component of these RHIM-dependent oligomers, with other proteins acting to control their formation. Together, these data show that RIPK3 dimerization, in the absence of receptor signaling, is sufficient to nucleate the formation of a RHIM-dependent necrosome. Further, necroptotic signaling from this complex is potentiated by the kinase activity of RIPK1 and inhibited by caspase-8 in a manner analogous to – but independent of – that observed during TNF-mediated necroptosis.
Oligomerization of RIPK3 is both necessary and sufficient to trigger necroptosis
Based on these data we hypothesized that RIPK3 oligomerization itself is key to its activation, and that RIPK1 and caspase-8 act by regulating the initiation and propagation of RIPK3 oligomers. Accordingly, we reasoned that chemically induced oligomerization (rather than dimerization) of RIPK3 should eliminate the ability of RIPK1 and caspase-8 to control this process. To achieve this, we created constructs in which two FV domains are attached to the C-terminus of RIPK3, creating what we refer to as RIPK3-2xFV, with the goal of promoting AP1-induced crosslinking and oligomerization independent of the RHIM domain (Figure 1a). Notably, the application AP1 to cells expressing RIPK3-2xFV resulted in a faster and more robust necroptotic response, to a degree comparable to that observed on dimerization of RIPK3-1xFV following caspase-8 inhibition or knockdown (Figure 3a compared with Figure 2a). Strikingly, although dimerization of a version of RIPK3 lacking a RHIM domain failed to induce any cell death response (Figure 1d), the addition of a second FV domain to create RIPK3ΔC-2xFV permitted robust necroptosis even in the absence of the RHIM domain (Figures 1a and 3b). As expected, cells expressing RHIM-less RIPK3ΔC-2xFV construct were non-responsive to TNF+zVAD (Supplementary Figure S1G.) Furthermore, addition of zVAD or Nec1 to cells expressing RIPK3-2xFV or RIPK3ΔC-2xFV did not alter magnitude or kinetics of cell death (Figures 3c and d). Consistent with a conclusion of chemically induced oligomerization, we observed large molecular weight RIPK3 complexes when cells expressing RIPK3-2xFV or RIPK3ΔC-2xFV were exposed to AP1 (Figure 3e). Interestingly, immunoprecipitation of RIPK3-2xFV following AP1 addition revealed interaction with RIPK1 and caspase-8, regardless of the presence of zVAD or Nec1 (Figure 3f). These data indicate that although RIPK1 and caspase-8 are recruited to the RIPK3 oligomers formed by this construct, enforced oligomerization of RIPK3 eliminates the ability of RIPK1 or caspase-8 to modulate RIPK3 activation. From this we can conclude that oligomerization of the RIPK3 kinase domain is both necessary and sufficient to trigger necroptosis, irrespective of the presence of the RHIM domain. Further, intrinsic control of this complex by caspase-8 and RIPK1 acts at, or upstream of, the formation of RIPK3 oligomers.
Chemically enforced RIPK3 oligomerization activates RIPK3 in the absence of the RHIM domain. (a–d) NIH-3T3 cells expressing RIPK3-2xFV (a and c) or RIPK3ΔC-2xFV (b and d) were treated as indicated, and cell death was assessed using an IncuCyte as described. (e) NIH-3T3 cells stably expressing indicated constructs were treated as indicated, lysed and necrosome complexes were covalently cross-linked using DSS. Resulting complexes were resolved by western blotting. (f) NIH-3T3 cells expressing the indicated constructs were treated as shown for 30 min, then lysed and subjected to immunoprecipitation using an antibody to the FLAG epitopes expressed on the RIPK3 constructs. Immune complexes were purified and resolved using TruBlot reagents to avoid aspecific signals from immunoglobulins, as described. Nec1 and zVAD were used at 30 and 50 μM, respectively, throughout
RHIM-dependent RIPK3 oligomerization is inhibited by the presence of RIPK1 protein
Our finding that ‘seeding’ of RHIM-dependent oligomers of RIPK3 via dimerization is inhibited by Nec1 (Figure 2c) implies that the kinase activity of RIPK1 drives the formation of RIPK3 oligomers; the recruitment of RIPK1 to RIPK3 dimers similarly supports the idea that RIPK1 may have a role in promoting the propagation of RIPK3 oligomers. To verify that our observations are not due to off-target effects of Nec1, we sought to confirm this finding by knocking down RIPK1 using siRNA (Supplementary Figure S2A). Unexpectedly, we found that although inhibition of RIPK1 with Nec1 blocked RIPK3 activation, RIPK1 knockdown significantly enhanced it (Figure 4a). This finding implies that the presence of the RIPK1 protein inhibits receptor-independent oligomerization of RIPK3, and that the inhibitory effects of Nec1 may rely on the scaffolding function of the chemically inhibited RIPK1 protein. Consistent with this idea, and with an on-target effect of Nec1, we found that the inhibitory effects of Nec1 on RIPK3 oligomerization were abrogated on knockdown of RIPK1 (Figure 4a compared with Figure 2c).
The presence of RIPK1 inhibits spontaneous RIPK3 oligomerization and necroptosis. (a) NIH-3T3 cells expressing RIPK3-1xFV were transfected with indicated siRNAs, then with 30 nM AP1 or 30 μM Nec1 72 h later. For scramble versus RIPK1 siRNA treated with AP1, *P=0.0024. (b) NIH-3T3 cells stably expressing DD-RIPK3 or DD-RIPK3K51A were treated with 1 μM Shield drug for indicated times, then lysed and resolved by western blotting. Jackson immortalized fibroblasts (Jax) expressing endogenous RIPK3 are included as a control. NIH-3T3 cells expressing DD-RIPK3 were transfected with indicated siRNAs, then treated 72 h later with 1 ng/ml recombinant TNF (c) or 1 μM Shield drug (d and e) and 30 μM Nec1 as indicated. c: *P<0.0001, d: *P<0.0001, e: *P=0.0001, **P<0.0001
These data imply that although the kinase activity of RIPK1 can potentiate RIPK3 oligomerization, RIPK1 is also required to exert intrinsic control of RIPK3 activation in the cytosol. We therefore reasoned that cells lacking RIPK1 should display reduced sensitivity to TNF-induced RIPK3 activation, but increased sensitivity to spontaneous activation of RIPK3. To directly test this idea, we fused RIPK3 to a destabilization domain (DD),33 creating a version of RIPK3 that is rapidly and constitutively degraded, but that accumulates in response to the DD-binding drug, referred to as Shield (Supplementary Figure S3A). We confirmed that the DD-RIPK3 fusion protein accumulated in response to Shield administration (Figure 4b), and that Shield pre-treatment increased the sensitivity of NIH-3T3 cells expressing this construct to TNF-induced necroptosis (Supplementary Figure S3B). Furthermore, RIPK3 accumulation was also sufficient to trigger spontaneous necrosome formation and limited cell death in the absence of exogenous TNF. Consistent with RIPK3 accumulation triggering spontaneous necrosome formation, this cell death was unaffected by the TNF-blocking reagent TNFR1-Fc (Supplementary Figure S3C), and could be abrogated by the K51A insertional mutation of the active site of DD-RIPK3 (Figure 4b and Supplementary Figure S3D).
We next used this system to evaluate RIPK1 as an intrinsic inhibitor of RIPK3 activation in the absence of receptor signaling. Consistent with canonical roles of caspase-8 and RIPK1 following TNFR1 ligation, knockdown of caspase-8 greatly sensitized cells expressing low levels of RIPK3 to TNF-induced cell death, while knockdown of RIPK1 did not (Figure 4c). However, when either caspase-8 or RIPK1 expression were silenced in the presence of Shield drug, the stabilization of RIPK3 was sufficient to support spontaneous, TNF-independent activation of DD-RIPK3 (Figure 4d). Importantly, addition of the RIPK1 inhibitor Nec1 to DD-RIPK3-expressing cells decreased cell death triggered by RIPK3 accumulation, and this effect was eliminated by RIPK1 siRNA-mediated knockdown, demonstrating that the effects of Nec1 are on-target and depend on the presence of the RIPK1 protein (Figure 4e).
To further explore these findings, we stably reconstituted murine embryonic fibroblast cells lacking both RIPK1 and RIPK3 (RIPK1/3 DKO mouse embryonic fibroblast (MEF)) with RIPK1 or a catalytically inactive RIPK1K45A mutant protein, and also expressed DD-RIPK3 in these cells (Figure 5a). Consistent with a role for RIPK1 in preventing RIPK3 activation at steady state, we were unable to achieve stable expression of DD-RIPK3 in RIPK1/3 DKO MEF cells not reconstituted with RIPK1 (not shown). Moreover, consistently we found that RIPK3 accumulation triggered spontaneous necroptosis to a notably greater degree in DKO MEF cells reconstituted with RIPK1, as compared with those expressing RIPK1K45A (Figure 5b). Nec1 inhibited RIPK3 activation and necroptosis in cells expressing RIPK1, but had no effect on cells expressing RIPK1K45A. These data indicate that although RIPK1 can drive receptor-induced RIPK3 activation and necroptosis, it also acts as an intrinsic suppressor of RIPK3 necrosome formation in the absence of receptor signaling. Furthermore, the RIPK1 inhibitor Nec1 potentiates this inhibitory function by creating an inactive form of RIPK1, an effect that is recapitulated by a catalytically inactive form of RIPK1. Genetic elimination and chemical inhibition of RIPK1 thereby have opposing effects.
Catalytically inactive RIPK1 mimics the suppressive effects of Nec-1 on spontaneous RIPK3 oligomerization and necroptosis. (a) RIPK1/RIPK3 DKO MEF cells were stably transduced with wild-type murine RIPK1 or RIPK1K45A, then stably transduced with DD-RIPK3. These cell lines were treated with Shield drug as indicated, then lysed and resolved by western blotting. Jax cells are included as a control for endogenous RIPK expression. (b) The cell lines depicted in a were treated with Shield drug in combination with Nec-1 as indicated. *P=0.0154, **P=0.0027. (c and d) A speculative model for the control of RIPK3 activation by RIPK1; RHIM domains are depicted in green, death domains in red and death effector domains in yellow. (c) When RIPK1 is activated by receptor signals, it binds to and phosphorylates RIPK3 via RHIM–RHIM interactions. This leads to RHIM-driven RIPK3 oligomerization, MLKL recruitment and necroptosis. (d) In the absence of receptor signals, spontaneous RIPK3 activation recruits RIPK1, which in turn recruits caspase-8/FLIP complexes. This leads to inhibition of RIPK3 oligomerization, possibly via cIAP-mediated degradation of the nascent RIPK3 complex
Discussion
In this study, we use inducible protein interaction systems to explore the mechanisms of RIPK3 activation. Our findings indicate that the formation of a RIPK3 dimer via a C-terminal dimerization domain, while not itself sufficient to activate RIPK3, is able to ‘seed’ an RHIM-dependent complex whose propagation leads to RIPK3 activation. Interestingly, we find that the stability of this complex, and by extension the activation of RIPK3 and necroptosis, is controlled by caspase-8 and RIPK1. Both caspase-8 and RIPK1 are recruited to the RIPK3 oligomer, and inhibition of caspase-8 potentiates RIPK3 oligomerization and necroptosis, while inhibition of RIPK1 inhibits these processes. Surprisingly, however, although siRNA-mediated knockdown of caspase-8 has a similar effect on its chemical inhibition, knockdown and inhibition of RIPK1 had opposing effects on RIPK3 oligomerization and activation. Although RIPK1 inhibition reduced RIPK3 activation, siRNA-mediated knockdown of RIPK1 notably potentiated it. From this, we can conclude that although the kinase activity of RIPK1 can contribute to RIPK3 activation, recruitment of RIPK1 is not required for – and indeed inhibits – the formation of RIPK3 oligomers. Consistent with this model, a recent study demonstrated that a knock-in mouse expressing catalytically inactive RIPK1D138N is viable, unlike RIPK1 knockout animals which die perinatally.34 This raises the intriguing possibility that the perinatal lethality of RIPK1-deficient animals may be the result of unchecked RIPK3 activity, possibly independently of receptor signaling.
In a companion to our study, Han and colleagues35 used similar induced-interaction systems to define the minimal complex necessary for necroptotic signaling. Although their results are largely compatible with ours, one key difference exists: they find that enforced RIPK3 dimerization, even in the absence of a RHIM domain, is sufficient to trigger RIPK3 autophosphorylation, MLKL activation and necroptosis. A likely explanation for this apparent discrepancy is that Han and colleagues appended dimerization domains adjacent the N-terminal kinase domain of RIPK3, while our study used C-terminal dimerization domains in an effort to mimic the action of the C-terminal RHIM domain. It is therefore likely that while forcing dimerization via the N terminus of RIPK3 leads to proximity-induced autophosphorylation and MLKL binding, a lack of structural constraint between the N-terminal kinase domain and the C-terminal RHIM means that C-terminal dimerization – as would occur with RHIM-RHIM interactions – does not.
These data point to a model in which RIPK3 dimerization leads to the exposure of the RHIM domain, allowing the recruitment of both RIPK1 and additional molecules of RIPK3 into the amyloid-like oligomer previously described.17 When RIPK1 is present in this structure, it mediates the recruitment of caspase-8 which, in concert with its paralog cFLIPL, promotes destabilization and inhibition of the growing necrosome via interactions between the C-terminal death domain of RIPK1 and the caspase-8 adapter Fas-associated protein with a death domain (FADD).25 Nec1 treatment promotes this inhibition by effectively creating a catalytically inactive form of RIPK1, which nonetheless is able to act as a molecular scaffold to recruit inhibitory caspase-8. The precise cleavage and phosphorylation events that govern necrosome assembly and stability remain poorly understood,34 but degradation of nascent RIPK3 complexes by the cIAPs has been shown to play a key role in the suppression of RIPK3 activation.18, 19, 36 We were unable to recover stable RIPK1/RIPK3 complexes following RIPK3 dimerization in the presence of Nec1, consistent with degradation of these complexes. Notably, however, we found that chemically enforced oligomerization potentiated RIPK3 activation and eliminated the ability of caspase-8 and RIPK1 to control this process, despite their recruitment to RIPK3 oligomers. Together, these findings are consistent with the idea that RIPK1-dependent recruitment of caspase-8 to nascent RIPK3 complexes targets them for degradation before RIPK3 oligomer formation. RIPK3 oligomerization may thus represent a ‘point of no return’ for necroptotic signaling (Figures 5c and d).
Although many of our findings were generated using inducible protein interaction systems, they have clear implications for RIPK3 signaling under physiological conditions. Consistent with structural studies of the RHIM domains17 as well as other amyloid-forming proteins, it is likely that the RHIM domains of RIPK3 proteins in the cytosol of healthy cells are somewhat ‘sticky,’ undergoing limited interactions in the absence of exogenous signals. Because of the self-propagating nature of these structures, it is important that cells have in place mechanisms to limit these interactions in the absence of pro-death signaling. Recruitment of the RIPK1 RHIM domain to RIPK3 oligomers, in the absence of other signals, may thereby recruit suppressive proteins to limit oligomer propagation. Understanding how RIPK1 is activated at the molecular level – that is, how recruitment to the TNFR1 complex and subsequent posttranslational modification potentiates the kinase activity of RIPK1 – will be important in understanding how RIPK1 makes the switch from inhibitor to activator of RIPK3 oligomerization during receptor signaling.
Materials and Methods
Constructs and cell lines
FLAG-RIPK3-FV chimeric proteins were constructed by cloning full-length murine RIPK3, catalytically dead RIPK3K51A, RIPK3T231,S232A, RIPK3 lacking the final 42 amino acids (RIPK3ΔC), or RIPK3 bearing a 4 amino acid substitution in the RHIM motif, VQIG→AAAA, (RIPK3ΔRHIM) upstream of either 1 or 2 copies of FKBP carrying the F36V mutation, herein called ‘FV’ domains. When two FV domains were used, the first copy contained silent mutations to prevent DNA recombination. These RIPK3-FV fusion proteins were cloned into pBabe-Puro retroviral vectors containing T2A ribosome-skipping sequences derived from porcine teschovirus-1 (EGRGSLLTCGDVEENPGP) upstream of eGFP.
These constructs were transduced into NIH-3T3 cells using standard protocols for helper-dependent retroviral transduction. Transduced cells were selected for 5 days in 1 μg/ml Puromycin, then grown to confluence and sorted twice for homogenous GFP expression. A minimum of two distinct, separately derived stable cell lines expressing each of these proteins was generated, and experiments presented are representative of results obtained with both cell lines. All cell lines were maintained in D-MEM (Fisher, Pittsburgh, PA, USA; SH30022FS) supplemented with 10% FCS (Sigma, St. Louis, MO, USA; 0926–500 Ml), 29.2 g/l glutamine (Fisher, SH3003402), 10 000 U/ml penicillin and 10 000 μg/ml streptomycin (Fisher, SV30010) and grown at 37 °C in 5% CO2.
Chimeric proteins composed of RIPK3 fused to the DD (described in Vanden-Verghe et al.31) were created via recombinant PCR, to produce a fragment composed of an N-terminal FLAG tag followed by the DD, then full-length murine RIPK3 or RIPK3K51A. These constructs were cloned into the pRRL lentiviral vector downstream of an MND promoter, and upstream of a T2A-GFP-T2A-Puromycin resistance cassette. RIPK1 or RIPK1K45A were expressed in pRRL vectors upstream of T2A-mCherry. E1A/Ras-transformed RIPK1/RIPK3 double-knockout MEF cells were first transduced with RIPK1 constructs, sorted for mCherry expression by FACS, then transduced with DD-RIPK3 and sorted for mCherry/GFP double positivity. NIH-3T3 cells transduced with DD-RIPK3 were selected in 2 μg/ml puromycin then sorted by FACS to ensure homogeneous expression. ‘Jax’ cells are a line of SV40 immortalized C57Bl/6 murine embryonic fibroblasts produced by the Jackson Laboratory (Bar Harbor, ME, USA).
Cell death assays
Cell death assays were carried out using a 2-color IncuCyte Zoom in-incubator imaging system (Essen Biosciences, Ann Arbor, MI, USA). Briefly, this system allows fully automated imaging of cells at set intervals in phase contrast as well as both red and green fluorescent channels. Cell death assays were carried out by treating cells with death-inducing compounds in 24-well tissue culture vessels (100 000 cells/well), in the presence of 100 nM of the cell-impermeable DNA-binding fluorescent dyes Sytox Green (Life Technologies, Grand Island, NY, USA; S7020) or Yoyo-3 (Y3606), which are excluded from healthy cells but rapidly enter dying cells on membrane permeabilization, in a manner analogous to propidium iodide. Resulting images were analyzed using the software package supplied with the IncuCyte imager, which allows precise analysis of the number of Sytox Green or Yoyo-3-positive cells present in each image. An example of cells undergoing necroptosis and the results of dead cell quantification are presented in Supplementary Movies 1 and 2. For each experiment, a minimum of three separate wells were treated with each experimental condition and a minimum of 4 image fields were assessed per well. Percent cell death was calculated by treating a minimum of three distinct wells in each experiment with 100 nM of the cell-permeable fluorescent dye Syto Green (S7559), which allows quantification of the total number of cells present in each field. Dead cell events acquired via Sytox Green or Yoyo-3 staining were divided by this total cell number to yield percent cell death at each time point. Error bars represent S.D. from the mean of a minimum of three independent wells. Statistical significance was calculated by two-way ANOVA using GraphPad Prism software. Each result depicted is representative of at least four distinct experiments, each of which contained at least three technical replicates.
Chemicals, compounds and siRNAs
The following compounds were used: recombinant murine TNF (Peprotech, Rocky Hill, NJ, USA; 315-01A) was used at 1 ng/ml unless otherwise specified; AP1 (now commercialized by Clontech as ‘B/B Homodimerizer,’ catalog number 635059) was dissolved in ethanol to a concentration of 100 μM, then diluted in culture media to a final concentration of 30 nM unless otherwise indicated. zVAD (SM Biochemicals, Anaheim, CA, USA; SMFMK001) was dissolved in DMSO to a concentration of 50 mM, then diluted to 50 μM in culture media; Nec-1 (Sigma, N9037-10MG) was dissolved in DMSO to a concentration of 30 mM, then diluted to 30 μM in culture media; TNFR-Fc (Fisher, 430-RI-050) was used at a final concentration of 200 ng/ml; shield drug was a kind gift of Dr. Tom Wandless and was used at a final concentration of 1 μM.
SiGenome SMARTpool siRNAs were purchased from Dharmacon/Fisher (Fisher Scientific, Pittsburgh, PA, USA). Pools targeting murine MLKL(M-061420-01), murine RIPK1 (M-040150-01) and murine caspase-8 (M-043044-01), as well as a non-targeting ‘scramble’ pool (D-001206-14), were used. Two microliters of a 50-μM stock of these siRNAs were delivered to cells in six-well format using Lipofectamine siRNA-Max reagent (Life Technologies, 13778150) according to the manufacturer’s instructions. Forty-eight hours later, these cells were re-plated into 24-well format for cell death assays, or collected for western blot analysis of knockdowns.
Antibodies and immunoprecipitation
The following antibodies were used: anti-caspase-8 (Enzo, Farmingdale, NY, USA; 1G12, ALX-804-447-C100), anti-RIPK1 (BD 610458), anti-RIPK3 (Imgenex, San Diego, CA, USA; IMG-5523-2), rabbit anti-FLAG (Abcam, Cambridge, MA, USA; AB1162), anti-actin (Millipore, Billerica, MA, USA; MAB1501), anti-FADD (Santa Cruz Biotechnology, Santa Cruz, CA, USA; M19). The anti-2A antibody was a kind gift from Dr. Dario Vignali. The anti-MLKL antibody was a kind gift from Dr. Warren Alexander.37 Secondary antibodies were purchased from Santa Cruz Biotechnology (mouse sc-2005, rat sc-2006 and rabbit sc-2313). These antibodies were used for western blotting of proteins separated using SDS-PAGE pre-cast gels (Invitrogen, Grand Island, NY, USA) by standard protocols. Detection was accomplished using either standard autoradiography film (Pierce, Rockford, IL, USA) or an electronic luminescence detection platform (ChemiDoc XRS+ System, 170-8265, Bio-Rad, Hercules, CA, USA).
Immunoprecipitation of necrosome complexes were carried out using rabbit anti-FLAG antibodies, and TruBlot IP reagents, now sold by Rockland Immunochemicals (Gilbertsville, PA, USA), to eliminate aspecific signals from immunoglobulin heavy and light chains. Briefly, cell lines were treated as indicated and lysed in NP40 buffer (30 mM Tris, 150 mM NaCl and 1% NP40) supplemented with protease inhibitors. Twenty micrograms of each sample were reserved for ‘input,’ while 500 μg of total protein from each sample were immunoprecipitated with 0.5 μg antibody conjugated to 30 μl TruBlot anti-Rabbit IgG beads. Immunocomplexes were washed four times in NP40 buffer, eluted by boiling, then run on western blotting. Western blottings were analyzed using standard primary antibodies, but TruBlot anti-Rabbit and anti-Mouse HRP-conjugated secondary antibodies were used. A standard anti-Rat-HRP secondary was used to detect caspase-8.
DSS crosslinking
A confluent monolayer of the indicated cells was incubated with the appropriate treatment for 30 min at 37 °C. Cells were lysed in a modified, Tris-free NP40 buffer (30 mM HEPES pH 7.4, 150 mM NaCl and 1% NP40) without protease inhibitors. Protein lysate was quantified according to standard BCA assay (Pierce) and 50 μg of each sample was treated with 0.1 mM DSS crosslinking agent (Thermo Scientific, Fisher Scientific, 21658) for 30 min at room temperature; the reaction was quenched by the addition of Tris-HCl pH 7.2 to a final concentration of 50 mM for 15 min. Crosslinked samples were analyzed by western blotting as described above, using the anti-2A primary antibody for detection.
Abbreviations
- AP1:
-
homodimerization drug
- cFLIPL:
-
cellular flice-like inhibitory protein, long isoform
- DAI:
-
DNA-dependent activator of interferon regulatory factor
- DD:
-
destabilization domain
- FADD:
-
Fas-associated protein with a death domain
- FKBP:
-
FK506-binding protein
- FV:
-
FKBPF36V point mutant
- MEF:
-
mouse embryonic fibroblast
- MLKL:
-
mixed-lineage kinase-like
- Nec1:
-
Necrostatin-1
- RHIM:
-
RIP homotypic interaction motif
- RIPK:
-
receptor-interacting protein kinase
- TNF:
-
tumor necrosis factor
- TNFR1:
-
TNF receptor-1
- TNFR-Fc:
-
TNFR1–immunoglobulin Fc fusion protein
- zVAD:
-
Z-val-ala-asp-(O-methlylated)-fluoromethylketone
References
Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G . Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714.
Vercammen D, Vandenabeele P, Beyaert R, Declercq W, Fiers W . Tumour necrosis factor-induced necrosis versus anti-Fas-induced apoptosis in L929 cells. Cytokine 1997; 9: 801–808.
Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321.
Lin Y, Choksi S, Shen HM, Yang QF, Hur GM, Kim YS et al. Tumor necrosis factor-induced nonapoptotic cell death requires receptor-interacting protein-mediated cellular reactive oxygen species accumulation. J Biol Chem 2004; 279: 10822–10828.
Cho YS, Challa S, Moquin D, Genga R, Ray TD . Guildford M, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123.
Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336.
He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 1100–1111.
Vanden Berghe T, Vanlangenakker N, Parthoens E, Deckers W, Devos M, Festjens N et al. Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features. Cell Death Differ 2010; 17: 922–930.
Thapa RJ, Nogusa S, Chen P, Maki JL, Lerro A, Andrake M et al. Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc Natl Acad Sci USA 2013; 110: E3109–E3118.
He S, Liang Y, Shao F, Wang X . Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc Natl Acad Sci USA 2011; 108: 20054–20059.
Upton JW, Kaiser WJ, Mocarski ES . DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 2012; 11: 290–297.
Upton JW, Kaiser WJ, Mocarski ES . Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 2010; 7: 302–313.
Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S et al. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat Immunol 2012; 13: 954–962.
Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 2011; 35: 908–918.
Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P . The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 1998; 8: 297–303.
Wang L, Du F, Wang X . TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703.
Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012; 150: 339–350.
Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M et al. cIAPs block ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 2011; 43: 449–463.
Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F et al. The ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43: 432–448.
Chen W, Zhou Z, Li L, Zhong CQ, Zheng X, Wu X et al. Diverse sequence determinants control human and mouse receptor interacting protein 3 (RIP3) and mixed lineage kinase domain-like (MLKL) interaction in necroptotic signaling. J Biol Chem 2013; 288: 16247–16261.
Wu J, Huang Z, Ren J, Zhang Z, He P, Li Y et al. Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res 2013; 23: 994–1006.
Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227.
Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA 2012; 109: 5322–5327.
Geserick P, Hupe M, Moulin M, Wong WW, Feoktistova M, Kellert B et al. Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol 2009; 187: 1037–1054.
Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C et al. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011; 471: 363–367.
Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011; 471: 368–372.
Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci USA 1998; 95: 10437–10442.
Oberst A, Pop C, Tremblay AG, Blais V, Denault JB, Salvesen GS et al. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J Biol Chem 2010; 285: 16632–16642.
Pop C, Oberst A, Drag M, Van Raam BJ, Riedl SJ, Green DR et al. FLIP(L) induces caspase 8 activity in the absence of interdomain caspase 8 cleavage and alters substrate specificity. Biochem J 2011; 433: 447–457.
Chang DW, Yang X . Activation of procaspases by FK506 binding protein-mediated oligomerization. Sci STKE 2003; 2003: PL1.
Vanden Berghe T, van Loo G, Saelens X, Van Gurp M, Brouckaert G, Kalai M et al. Differential signaling to apoptotic and necrotic cell death by Fas-associated death domain protein FADD. J Biol Chem 2004; 279: 7925–7933.
Dillon CP, Oberst A, Weinlich R, Janke LJ, Kang TB, Ben-Moshe T et al. Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep 2012; 1: 401–407.
Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ . A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 2006; 126: 995–1004.
Newton K, Dugger DL, Wickliffe KE, Kapoor N, de Almagro MC, Vucic D et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 2014; 343: 1357–1360.
Wu X-N, Yang Z-H, Wang X-K, Zhang Y, Wan H, Song Y et al. Distinct roles of RIP1–RIP3 hetero- and RIP3–RIP3 homo-interaction in mediating necroptosis. Cell Death Differ 2014 e-pub ahead of print 6 June 2014; doi:10.1038/cdd.2014.77.
Feoktistova M, Geserick P, Panayotova-Dimitrova D, Leverkus M . Pick your poison: the Ripoptosome, a cell death platform regulating apoptosis and necroptosis. Cell Cycle 2012; 11: 460–467.
Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 2013; 39: 443–453.
Acknowledgements
We thank Dr. Dan Stetson and Dr. Ram Savan, and their groups, for moral and material support. We also thank Dr. Stetson and Dr. David Rawlings for providing pRRL lentiviral constructs, Drs Dario Vignali and Warren Alexander for providing antibodies and Dr. Christopher Dillon for preparing RIPK1/3 DKO MEFs. We also thank Dr. Jiahuai Han for helpful discussions and input. This work was supported by NIH grant RAI108685A, Royalty Research Fund grant 65-0062 and UW Diabetes Research Center grant P30 DK017047, to AO. SO is supported by an NIH training grant 1T32AI106677-01 to the Department of Immunology of the University of Washington.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Orozco, S., Yatim, N., Werner, M. et al. RIPK1 both positively and negatively regulates RIPK3 oligomerization and necroptosis. Cell Death Differ 21, 1511–1521 (2014). https://doi.org/10.1038/cdd.2014.76
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.76
This article is cited by
-
Immunogenic cell death in cancer: targeting necroptosis to induce antitumour immunity
Nature Reviews Cancer (2024)
-
Repurposing of Ibrutinib and Quizartinib as potent inhibitors of necroptosis
Communications Biology (2023)
-
PRMT5-mediated regulatory arginine methylation of RIPK3
Cell Death Discovery (2023)
-
OASL phase condensation induces amyloid-like fibrillation of RIPK3 to promote virus-induced necroptosis
Nature Cell Biology (2023)
-
HSPA8 acts as an amyloidase to suppress necroptosis by inhibiting and reversing functional amyloid formation
Cell Research (2023)