Abstract
Although the Myc transcription factor has been shown necessary for the oncogenic function of Ras, the contribution of Ras pathway signaling to the oncogenic function of Myc remains unresolved. We report the novel findings that Myc alone induced Ras/Mapk pathway signaling, and increased signaling following growth factor stimulation. Deletion of the scaffold protein kinase suppressor of Ras 1 (Ksr1) attenuated signaling through the Ras/Mapk pathway, including activation following Myc induction. B cells that lacked Ksr1 exhibited reduced proliferation and increased cytokine deprivation-induced apoptosis. Overexpression of Myc rescued the proliferation defect of Ksr1-null B cells, but loss of Ksr1 increased sensitivity of B cells to Myc-induced apoptosis. Notably, there was a significant delay in lymphoma development in Ksr1-null mice overexpressing Myc in B cells (Eμ-myc transgenic mice). There was an elevated frequency of p53 inactivation, indicative of increased selective pressure to bypass the p53 tumor suppressor pathway, in Ksr1-null Eμ-myc lymphomas. Therefore, loss of Ksr1 inhibits Ras/Mapk pathway signaling leading to increased Myc-induced B-cell apoptosis, and this results in reduced B-cell transformation and lymphoma development. Our data indicate that suppression of Myc-induced Ras/Mapk pathway signaling significantly impairs Myc oncogenic function. These results fill a significant gap in knowledge about Myc and should open new avenues of therapeutic intervention for Myc-overexpressing malignancies.
Similar content being viewed by others
Main
The c-Myc transcription factor is an oncoprotein that is frequently overexpressed in hematological malignancies through translocations, amplifications, and other less characterized mechanisms. Overexpression of Myc, which drives cell cycle progression, is a well-characterized initiating step in human Burkitt's lymphoma, a non-Hodgkin's B-cell lymphoma. The role of Myc in non-Hodgkin's B-cell lymphoma was established in part by the Eμ-myc transgenic mouse model that overexpresses Myc in B-cells and develops pre-B/B-cell lymphoma.1 Overexpression of Myc in primary B cells triggers activation of the Arf-Mdm2-p53 tumor suppressor pathway, which leads to apoptosis. Specifically, Myc induces Arf, which inhibits Mdm2, causing p53 activation and B-cell apoptosis. Myc-overexpressing B cells that acquire mutations that inhibit apoptosis survive and can be transformed. This is exemplified in that 80% of the lymphomas that arise in Eμ-myc mice have deleted Arf, overexpress Mdm2, or have mutated or deleted p53, all of which inhibit Myc-induced apoptosis.2 Similar observations have been made in human lymphomas.3, 4, 5 Therefore, whereas Myc overexpression induces B-cell transformation in vivo, it requires B cells to gain resistance to apoptosis induced by the hyperproliferative signals from Myc for this to occur.
The Ras-Raf-Mek-Mapk/Erk (Ras/Mapk) signaling pathway is essential for cell growth, differentiation, and cell survival.6 Kinase suppressor of Ras 1 (Ksr1) was first identified as a positive modulator of Ras signaling in genetic screens of Drosophila and C. elegans.7 Subsequent studies have shown Ksr1 functions as a scaffold protein that can bind and facilitate signaling of multiple components of the Ras/Mapk signaling pathway.6 Deletion of Ksr1 results in decreased Mapk-Erk1/2 signaling.8 In lymphocytes, loss of Ksr1 inhibits T-cell activation, but does not influence T- or B-cell development.8 Consistent with its scaffolding function in the Ras/Mapk signaling pathway, Ksr1 overexpression or loss of expression inhibits oncogenic Ras (RasV12)-induced fibroblast transformation in vitro.9, 10 In vivo, deletion of Ksr1 inhibits Ras signaling-dependent tumor development in the mammary gland and skin.8, 11 However, there are incongruous data that Ksr1 can promote fibroblast proliferation,12 but also that deletion of Ksr1 increases spontaneous immortalization and RasV12-induced proliferation of fibroblasts.10 Therefore, Ksr1 has a critical function in modulating Ras pathway signaling, but its role in the cellular processes linked to transformation and in tumorigenesis remain incompletely resolved.
Oncogenic Ras mutations have been identified in leukemias and lymphomas.13, 14, 15 Ras mutations have also been reported in pre-B/B-cell lymphomas arising in Eμ-myc transgenic mice.16 Over 20 years ago, it was shown that co-expression of v-H-Ras and v-Myc transforms pre-B cells,17 and that v-H-Ras accelerates B-cell lymphomagenesis in Eμ-myc transgenic mice.18 Despite these findings, it remains unclear whether the endogenous Ras signaling cascade is necessary for or contributes to Myc-induced B-cell tumorigenesis. Crosses with Eμ-myc mice and Ksr1-null mice, which are disabled in their ability to signal properly through the Ras/Mapk pathway,6 reveal the importance of this cascade for Myc-mediated B-cell transformation. We show that Myc induction activates Mapk/Erk signaling, and that this response is attenuated in cells lacking Ksr1. Loss of Ksr1 sensitized B cells to Myc-induced apoptosis and significantly delayed Myc-mediated B-cell lymphomagenesis. Lymphomas that did emerge in Ksr1−/− Eμ-myc mice had an increased frequency of p53 inactivation. Our results indicate that the Ras pathway cooperates with Myc in oncogenic transformation of B cells by providing anti-apoptotic signaling. Therefore, modulators of Ras signaling, such as Ksr1, may prove to be efficacious therapeutic targets in Myc-driven cancers.
Results
Ksr1 facilitates Myc-induced Ras activity
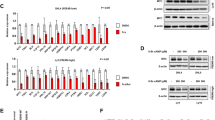
Despite the well-known synergy of Myc overexpression and oncogenic Ras in cellular transformation and a recent report that Myc is necessary for oncogenic Ras-induced lung cancer,19 the contribution of endogenous Ras signaling to the oncogenic function of Myc remains uncharacterized. Therefore, we investigated whether Myc could influence Ras signaling through the Ras-Raf-Mek-Mapk/Erk pathway. Wild-type mouse embryonic fibroblasts (MEFs) were infected with a bicistronic retrovirus encoding MycER, a 4-hydroxytamoxifen (4-OHT)-inducible form of Myc,20 and green fluorescent protein (GFP). GFP-positive MEFs treated with 4-OHT to activate MycER exhibited significantly increased basal Mek1/2 (4.4-fold) and Erk1/2 (7.2-fold) phosphorylation compared with ethanol vehicle control-treated MEFs (Figure 1a). Mek1/2 and Erk1/2 are downstream kinases activated by Ras signaling, and a measure of Ras activation. Ras is activated by growth factor receptor ligation leading to receptor tyrosine kinase activation.6 Therefore, as an additional test of the influence Myc has on Ras pathway activation, murine fibroblasts expressing MycER were pre-treated with 4-OHT or ethanol control, removed from growth factors, and then subsequently treated with serum or epidermal growth factor (EGF). Activation of MycER resulted in increased and sustained Ras/Mapk activation in response to both serum and EGF relative to vehicle control (Figure 1b). These data show that Myc can enhance not only basal, but also receptor tyrosine kinase-induced Ras/Mapk pathway signaling.
Myc activation enhances basal and receptor tyrosine kinase-stimulated Ras/Mapk activity. (a) Wild-type MEFs expressing MycER were treated with 4-OHT to activate Myc or ethanol (EtOH) vehicle control. Mek1/2 phosphorylation and Erk1/2 phosphorylation, as measures of Mapk activity, were assessed by western blot. (b) Murine fibroblasts (NIH3T3 cells) expressing MycER were treated with 4-OHT or EtOH control, washed with PBS, and then treated with complete media containing 10% serum (top panel) or serum-free media containing 100 ng/ml EGF (bottom panel). Erk1/2 phosphorylation was assessed by western blot. (c) MEFs from Ksr1+/+ and Ksr1−/− embryos expressing MycER were treated with 4-OHT to activate MycER, and basal Erk1/2 phosphorylation was assessed by western blot. Relative levels of phosphorylated Erk1/2 and Mek1/2 are indicated
Previous studies have demonstrated that Ksr1 acts as a scaffolding protein that positively regulates Ras pathway signaling and cooperates with oncogenic Ras in transformation.7 To test whether Ksr1 contributes to Ras/Mapk pathway signaling induced by Myc, we evaluated MEFs lacking Ksr1. Addition of 4-OHT to MycER-expressing Ksr1−/− MEFs resulted in a 3.1-fold reduction in Erk1/2 phosporylation compared with MycER-induced Erk1/2 phosphorylation in Ksr1+/+ MEFs (Figure 1c). Therefore, loss of Ksr1 attenuates Myc activation of the Ras/Mapk pathway.
Lack of Ksr1 reduces proliferation and increases apoptosis of pre-B cells
As overexpression of Myc is thought to be an initiating event in the development of some B-cell lymphomas, we sought to determine the contribution of Ras signaling to Myc-induced B-cell lymphomagenesis by utilizing Ksr1-deficient mice. We first investigated the role of Ksr1 in B-cells in the absence of Myc overexpression. In agreement with Nguyen et al.,8 analysis of bone marrow from mice lacking one or both alleles of Ksr1 revealed no significant defects in B-cell development (data not shown). However, Ksr1−/− pre-B cells emerged from cultured primary bone marrow more slowly than littermate-matched Ksr1+/+ pre-B cells (Figure 2a). Moreover, in repeated experiments, Ksr1−/− pre-B cells exhibited consistently reduced population doublings in culture compared with Ksr1+/+ pre-B cells (Figure 2a, representative experiment). Population doublings of Ksr1+/− pre-B cells were similar to Ksr1+/+ pre-B cells (data not shown). The reduction in cell growth of Ksr1−/− pre-B cells was confirmed by cell counting and MTT assay relative to the same starting number of cells over 6 days (Figures 2b and c). To determine if the difference in cell expansion was due to a difference in proliferation, and to specifically evaluate the percentage of cells in S phase, BrdU incorporation was assessed. Ksr1−/− pre-B cells exhibited significantly reduced BrdU incorporation (reduction of 19.0%) relative to Ksr1+/+ pre-B cells (Figure 2d, P<0.0001). Together, these data indicate a lack of Ksr1 results in reduced pre-B cell proliferation.
Pre-B cells lacking Ksr1 exhibit reduced proliferation and increased apoptosis. (a) Bone marrow from Ksr1+/+, Ksr1+/−, and Ksr1−/− littermates was placed into media containing IL-7 on day 0. Viable cells were counted at intervals by Trypan Blue Dye exclusion assay. Population doublings were calculated. A representative experiment is shown; similar results were obtained with bone marrow from littermates from different parents. (b and c) Equal numbers of Ksr1+/+ and Ksr1−/− pre-B cells were plated in triplicate and the total number of viable cells was assessed by Trypan Blue Dye exclusion (b) or MTT assay (c) for 6 days. (d) Ksr1+/+ and Ksr1−/− pre-B cells were pulsed with BrdU, and incorporation of BrdU was determined by flow cytometry. Data are expressed as average BrdU incorporation relative to Ksr1+/+ cells (n=4 of each); P<0.0001. (e) Equal numbers of Ksr1+/+ and Ksr1−/− pre-B cells were cultured in media lacking IL-7 beginning at day 0. Viability was monitored for 3 days by Trypan Blue Dye exclusion assay. Percent viability at each time is expressed relative to the viability at time 0
In addition to reduced proliferation, we also investigated whether apoptosis may have also contributed to the decreased cell numbers observed with Ksr1−/− pre-B cells. Analysis of cell viability of Ksr1−/− and Ksr1+/+ pre-B cells revealed consistently more dead and apoptotic cells in the Ksr1−/− cultures (data not shown). We also observed that Ksr1−/− pre-B cells were significantly more sensitive to IL-7 deprivation-induced apoptosis than wild-type Ksr1+/+ pre-B cells (Figure 2e). Therefore, loss of Ksr1 leads to both decreased proliferation and increased spontaneous and growth factor deprivation-induced apoptosis.
Loss of Ksr1 increases Myc-induced apoptosis
Given that Myc induces Ras/Mapk pathway signaling and that this is inhibited by loss of Ksr1 (Figure 1), we evaluated the biological consequences of suppressing Ras pathway signaling in B cells overexpressing Myc. We crossed Ksr1-deficient mice to Eμ-myc transgenic mice that overexpress Myc in the B-cell compartment and subsequently develop spontaneous pre-B/B-cell lymphoma.1 Prior to lymphoma development, B-cell development was evaluated in bone marrow of Ksr1−/− Eμ-myc mice. There were no detectable differences in B-cell development between Ksr1−/− Eμ-myc, Ksr1+/− Eμ-myc, and Ksr1+/+ Eμ-myc littermates (data not shown). However, in repeated experiments, Ksr1−/− Eμ-myc and Ksr1+/− Eμ-myc pre-B cells derived from cultured bone marrow harvested prior to lymphoma development consistently exhibited reduced population doublings relative to littermate control Ksr1+/+ Eμ-myc pre-B cells (Figure 3a, representative experiment). Moreover, once pre-B cells emerged from the cultures (days 10–12), the number of viable pre-B cells of each genotype was determined each day for 7 days by Trypan Blue Dye. Ksr1−/− Eμ-myc pre-B cells consistently had decreased numbers of viable pre-B cells compared with Ksr1+/+ Eμ-myc pre-B cells (Figure 3b). The number of viable Ksr1+/− Eμ-myc pre-B cells was intermediate. An additional measure of cell growth by MTT assay further confirmed the decreased viability of Ksr1−/− Eμ-myc pre-B cells relative to Ksr1+/+ Eμ-myc pre-B cell cultures (Figure 3c). Given that Erk activity can influence Myc protein stability,21 which could account for the differences in cell number we detected, we assessed Myc levels in Eμ-myc pre-B cells. There were similar levels of Myc protein in all three genotypes of Eμ-myc pre-B cells (Figure 3d). Therefore, the differences in the rate of pre-B cell growth between the Ksr1−/− Eμ-myc and Ksr1+/+ Eμ-myc pre-B cells are not due to differences in Myc expression.
Reduced growth of Ksr1 deficient Eμ-myc pre-B cells. (a) Bone marrow from littermates of the indicated genotype was placed into culture on day 0. Viable cells were counted by Trypan Blue Dye exclusion at intervals, and population doublings were calculated. A representative experiment is shown; similar results were obtained with bone marrow from littermates from different parents. (b and c) Equal numbers of Ksr1+/+ Eμ-myc, Ksr1+/− Eμ-myc, and Ksr1−/− Eμ-myc pre-B cells were placed in culture and viability was assessed by Trypan Blue Dye exclusion (b) and MTT assay (c). (d) Myc and β-actin protein levels in littermate-matched pre-B cells were determined by western blot
We postulated that differences in both proliferation and apoptosis would account for the decreased numbers of viable Ksr1−/− Eμ-myc pre-B cells. Surprisingly, there was no significant difference in the cell cycle profiles or the percentage of cells in S phase (BrdU incorporation) between Ksr1−/− Eμ-myc and Ksr1+/+ Eμ-myc pre-B cells (Figure 4a, P=0.97, and data not shown). However, there was a consistently elevated level of non-viable and apoptotic Ksr1−/− Eμ-myc pre-B cells relative to Ksr1+/+ Eμ-myc pre-B cells in culture (Figure 4b, P=0.0064). To specifically examine the contribution of Ksr1 to Myc-induced apoptosis, pre-B cells from littermate-matched Ksr1+/+ and Ksr1−/− mice (not Eμ-myc) were infected with a bicistronic retrovirus encoding MycER and GFP. GFP-positive pre-B cells of both genotypes expressed similar levels of MycER protein (Figure 4c). Following activation of MycER with 4-OHT, Ksr1−/− MycER pre-B cells exhibited markedly increased apoptotic cell death relative to Ksr1+/+ MycER pre-B cells (Figure 4c). The decreasing viability of the Ksr1−/− MycER pre-B cells was associated with a significantly increasing percentage of cells with sub-G1 DNA content over 12 hours of MycER activation (Figure 4d). Thus, loss of Ksr1 did not affect Myc-driven proliferation, but did increase the sensitivity of pre-B cells to Myc-induced apoptosis. Collectively, these data indicate that Ksr1 expression is protective against Myc-induced apoptosis.
Enhanced Myc-induced apoptosis in Ksr1−/− pre-B cells. (a) BrdU incorporation in Ksr1+/+ Eμ-myc and Ksr1−/− Eμ-myc pre-B cells was measured by flow cytometry (P=0.97, t-test.) (b) Steady-state levels of dead and dying cells in cultures of Eμ-myc pre-B cells of the indicated genotype were measured by Trypan Blue Dye. Data are relative to the Ksr1+/+ Eμ-myc cultures; *P=0.0249, **P=0.0064, t-test. (c) MycER-expressing Ksr1+/+ and Ksr1−/− pre-B cells were treated with 4-OHT to activate MycER at time 0. Viability was assessed by Trypan Blue Dye exclusion assay. Percent viability at each time is expressed relative to the initial viability at time 0; *P=0.0242, **P=0.0122, ***P=0.0028, t-test. Inset, MycER protein levels were examined by western blot with β-actin as a loading control. (d) DNA in MycER-expressing pre-B cells was stained with propidium iodide at intervals after treatment with 4-OHT and sub-G1 DNA content, as a measure of apoptosis, was analyzed by flow cytometry; *P=0.0175, **P=0.0267, t-test
Ksr1 deficiency inhibits Myc-driven B-cell lymphoma development
To assess the overall effect of reduced Ras/Mapk pathway signaling on Myc-induced B-cell lymphoma development, we generated a cohort of Eμ-myc transgenic mice deficient in Ksr1 and littermate controls. Ksr1−/− Eμ-myc mice demonstrated a significant delay in lymphoma development compared with Ksr1+/+ Eμ-myc littermates (Figure 5, P=0.0365 log-rank test). At 300 days, ∼30% of the Ksr1−/− Eμ-myc mice were alive, whereas only ∼10% of the Ksr1+/+ Eμ-myc mice were alive. Moreover, the mean survival of Ksr1+/+ Eμ-myc mice (95 days) was significantly lower than Ksr1−/− Eμ-myc mice (138 days). Ksr1+/− Eμ-myc mice exhibited an intermediate onset of lymphoma development and survival (122 days mean survival), but survival was not statistically different from Ksr1+/+ Eμ-myc mice (P=0.1752, log-rank test). The B-cell lymphomas arising in all three genotypes of mice were typical pre-B/B-cell lymphomas that emerge in wild-type Eμ-myc mice (data not shown). These data indicate that lack of Ksr1 significantly delays Myc-driven lymphoma development, but does not alter the type of B-cell malignancy that emerges.
Increased frequency of p53 inactivation in Ksr1−/− Eμ-myc lymphomas
The Arf-Mdm2-p53 tumor suppressor pathway is directly inactivated in at least 80% of lymphomas that emerge in Eμ-myc mice.2 To examine the effect of reduced Ras/Mapk pathway signaling from loss of Ksr1 has on the Arf-Mdm2-p53 pathway in Myc-induced lymphomas, Arf, Mdm2, and p53 status were evaluated in Ksr1−/− Eμ-myc lymphomas. Biallelic deletion of Arf was detected by Southern blot in 6 of the 28 (21.4%) Ksr1−/− Eμ-myc lymphomas analyzed (Figure 6a). This frequency is comparable to that previously observed in wild-type Eμ-myc lymphomas (24%).2 Missense mutations in p53 can be detected by western blot, as these mutations typically lead to p53 protein accumulation with a concomitant increase in Arf protein.2 Eight of the 28 lymphomas analyzed had increased p53 protein, with mutations in the DNA-binding domain of p53 being the most common mutation detected (Figure 6b and data not shown). As p53 mutation or deletion leads to Arf overexpression,2, 22 p53 was sequenced and Southern blots were performed on genomic DNA from Ksr1−/− Eμ-myc lymphomas that did not exhibit p53 overexpression but did overexpress Arf (Figure 6b). This analysis revealed that three lymphomas contained nonsense mutations in p53 leading to truncation of p53, but no deletions of p53 were detected in Ksr1−/− Eμ-myc lymphomas (data not shown). Combined, the frequency of p53 mutations in Ksr1−/− Eμ-myc lymphomas was 39.3% (11 of 28), which is higher than what has been typically observed in wild-type Eμ-myc lymphomas2, 23 and littermate control Ksr1+/+ Eμ-myc lymphomas in this study (2 of 10; 20%). p53 can also be inactivated by Mdm2 overexpression. Of the 28 Ksr1−/− Eμ-myc lymphomas analyzed, 18 (64.3%) exhibited overexpression of at least one of the three Mdm2 isoforms. This frequency is also elevated relative to that previously reported for wild-type Eμ-myc lymphomas (48%) as well as littermate control Ksr1+/+ Eμ-myc lymphomas analyzed in parallel in this experiment (5 of 10; 50%). Collectively, our results indicate that lack of Ksr1 does not affect the frequency of Arf deletion, but does increase the incidence of Mdm2 overexpression and p53 mutations in Eμ-myc lymphomas. These data support a model in which lack of Ksr1 increases the selective pressure to overcome p53 tumor suppressor activity, resulting in an increased frequency of Mdm2 overexpression and p53 mutations.
Analysis of p53, Arf, and Mdm2 status in Ksr1−/− Eμ-myc lymphomas. (a) Southern blot for Arf in lymphomas from Ksr−/− Eμ-myc transgenic mice where Arf was not detectable by western blot. Lymphomas that had biallelic Arf deletion are denoted by an asterisk (*). Control DNA for wild-type (WT) Arf loci and biallelic Arf deletion (del) are indicated. (b) Protein lysates of lymphomas from Ksr1−/− Eμ-myc transgenic mice were western blotted for p53, Arf, Mdm2, and as a loading control, β-actin. Lysates from lymphomas that overexpress Mdm2 and Arf or mutant p53 and p53-null MEFs are controls. An asterisk (*) denotes lymphomas with mutant p53 as determined by sequencing. The percentage of lymphomas that contained the indicated alterations is shown
Discussion
Previous studies have evaluated the contribution of Ras to Myc-induced B-cell malignancy by forced expression of oncogenic Ras and Myc, which transforms B cells in vitro and in vivo.17, 18 Screening for alterations in Ras and/or proteins in the Ras pathway in leukemias and lymphomas has also been done. Both gain-of-function mutations in Ras isoforms (H-Ras, K-Ras, or N-Ras) and loss-of-function mutations that directly activate Ras/MAPK signaling have been identified in human and murine leukemias and lymphomas, many of which overexpress Myc.14, 16, 24 In addition, analysis of human B-cell malignancies showed a frequent increase in the copy number of genomic regions that contain Ras pathway components.25 However, none of the studies examined whether endogenous Ras pathway signaling contributes to Myc-induced lymphoma development or how Ras pathway signaling impacts Myc-driven tumorigenesis. Utilizing primary cells and mouse models, we have shown that deletion of Ksr1, an established positive regulator of Ras signaling, leads to inhibition of Myc-induced Ras/Mapk pathway signaling, increased Myc-initiated apoptosis, and a significant delay in Myc-mediated lymphomagenesis. Our data significantly extend previous knowledge on Myc-induced transformation and highlight the critical function of endogenous Ras/Mapk pathway signaling in Myc oncogenic function.
Apoptosis inhibits hyperproliferative signals from oncogenes, such as Myc, and thus limits the development of hematopoietic malignancies. Acquiring resistance to Myc-induced apoptosis is a requisite step in Myc-mediated lymphomagenesis.2 B-cells that are resistant to Myc-induced apoptosis, such as those that lack Arf, p53, or pro-apoptotic Bcl2 family members, and those that overexpress Mdm2 or anti-apoptotic Bcl2 family members exhibit accelerated Myc-mediated lymphomagenesis.2, 23, 26, 27, 28 Increased sensitivity of B cells to Myc-induced apoptosis, such as that conferred with Mdm2 heterozygosity, inhibits lymphoma development driven by Myc.23 As the Ras pathway can facilitate signaling that leads to cell growth as well as cell death, and Ksr1 inactivation is reported to lead to both increased and decreased sensitivity to p53-mediated apoptosis,29, 30 it was unclear whether or how suppression of this pathway by deletion of Ksr1 was going to impact the ability of Myc to transform B-cells. Our data showed that although loss of Ksr1 led to reduced proliferation and increased apoptosis of B-cells, in the presence of Myc overexpression proliferation rates were analogous, but apoptosis was increased. These results suggest that Myc overexpression compensated for decreased Ras/Mapk pathway signaling in proliferation, but normal Ras/Mapk pathway signaling was required to mediate the cell survival signal. The increased frequency of p53 inactivation in Ksr1−/− Eμ-myc lymphomas we detected is also supportive of this conclusion. However, the precise mechanism for the increased sensitivity of Ksr1−/− pre-B cells to Myc-induced apoptosis remains unresolved. Evaluation of anti- and pro-apoptotic Bcl2 family members, particularly ones whose expression are known to be regulated by Myc (e.g., Bcl2, BclxL, and Bim31, 32, 33), in pre-cancerous Eμ-myc B cells did not reveal differences in expression of these proteins in the absence of Ksr1 (data not shown). Therefore, Ksr1 expression decreases the sensitivity of B cells to Myc-induced apoptosis by promoting Ras/Mapk pathway signaling, and this appears to be independent of the effects Myc has on Bcl2 family members.
Our results reveal a possible achilles’ heel in tumors that rely on Myc for growth and survival. Studies from multiple labs have established that loss or inhibition of Myc results in regression of many Myc-overexpressing tumors.34 It has recently been appreciated that Myc function is also critical for the maintenance and survival of mutant Ras-driven lung tumors, as inhibition of Myc led to regression of lung tumors that overexpressed K-RasG12D.19 It has also been hypothesized from studies using oncogenic Ras and Myc overexpression that targeting both Myc and Ras may be beneficial for treating lung cancers and lymphoma.35 However, Myc to-date has not been able to be targeted therapeutically in patients. Our results establish the critical function of the endogenous Ras/Mapk signaling pathway in Myc-induced lymphoma development and B-cell survival, and provide impetus to test inhibitors of Ras/Mapk pathway signaling in established Myc-overexpressing lymphomas. Previous reports have shown that Ksr1 expression appears important for tumor maintenance, although Myc levels were not investigated in these studies. Specifically, KSR1 antisense inhibited the proliferation of epidermoid carcinoma cells and the growth of pancreatic and lung tumor cells in xenografts.36 Furthermore, knockdown of KSR1 sensitized endometrial carcinoma cells to death receptor-mediated apoptosis.37 Although deletion of Mek1 inhibits tumor formation in the skin,38 it was shown that shRNA knockdown of Mek1 in murine hematopoietic stem cells facilitated Myc-induced tumor development.39 However, the decrease in survival of the mice with Mek1 knockdown was not reported to be statistically significant, but may reflect different requirements of Mek1 signaling in stem cells. Recently, a pharmacological MEK inhibitor was shown to kill human diffuse large B-cell lymphoma cells, which frequently overexpress Myc.40 Thus, a likely fruitful avenue of investigation will be to determine whether Ras/Mapk pathway signaling is critical for the maintenance and survival of lymphomas and the many other cancers that overexpress Myc.
Materials and Methods
Mice and phenotyping
Congenic C57Bl/6 Eμ-myc transgenic mice were made by Dr Alan Harris (Walter & Eliza Hall Institute, Melbourne, Australia)1 and obtained from Dr Charles Sidman (University of Cincinnati, Cincinnati, OH, USA). Ksr1+/− mice were made by Dr Jeffrey Stock (Pfizer, New York, NY, USA)8 and obtained from Dr. Robert Lewis (University of Nebraska Medical Center). Ksr1+/− mice (backcrossed onto C57Bl/6 at least 10 generations) were mated to male Eμ-myc transgenic mice to generate F1's. The F1's were then crossed to generate F2's for analysis. The transgene was always inherited through the male. Mice were carefully followed and euthanized when lymphomas developed. Statistical significance was determined by log-rank test. All experiments with mice were approved by the Vanderbilt Institutional Animal Care and Use Committee (IACUC) and followed all federal and state rules and regulations. All phenotyping procedures are previously described.23 Antibodies for phenotypic analyses of bone marrow, spleens, and lymphomas were from Southern Biotechnology (Birmingham, AL, USA) and BDPharMingen (San Diego, CA, USA).
Culture and infection of primary pre-B cells and fibroblasts
Primary pre-B cell (CD43−, B220+, CD19+, IgM−) cultures were generated from bone marrow of 4–6-week-old mice as previously described.2 For pre-B cell retroviral infections, bicistroinic retrovirus encoding MSCV-MycER-IRES-GFP was generated and used to infect pre-B cells as previously described.23 GFP-positive pre-B cells were isolated by fluorescence-activated cell sorting (FACS) and expanded for analysis. MEFs were generated from Ksr1+/+ and Ksr1−/− embryos from pregnant Ksr+/− females. MEFs and NIH3T3 cells were similarly infected with MSCV-MycER-IRES-GFP retrovirus, and GFP expression was evident in >95% of the MEFs.
Measurement of Ras/Mapk activity
In six-well plates, sub-confluent MycER-expressing fibroblasts were washed with PBS to starve them of serum following 24 hours of pre-treatment with 4-OHT or ethanol (ETOH) vehicle control. Media containing 10% serum or serum-free media supplemented with 100 ng/ml EGF (PHG0313, Invitrogen, Camarillo, CA, USA) was then added to the fibroblasts. Cells were then washed with cold PBS at intervals, lysed, and proteins western blotted as described below. For the basal Ras/Mapk activity experiments, fibroblasts in 10% serum expressing MycER were treated for 24 hours with EtOH vehicle control or 4-OHT and then lysed (see below).
Viability, apoptosis, proliferation, and cell cycle analysis
Viability of pre-B cells following explantation of bone marrow into IL-7-containing medium, after addition of 4-OHT (Sigma, St Louis, MO, USA) to culture media of MycER-expressing pre-B cells, or after IL-7 deprivation was determined at intervals by Trypan Blue Dye exclusion and MTT assay as previously described.23 Population doublings of bone marrow cultures were determined by counting cells excluding Trypan Blue Dye at intervals over 25–60 days. Proliferation was examined by BrdU incorporation as previously described.27 Quantification of fragmented (Sub-G1) DNA was determined following staining DNA with propidium iodide (PI). Fluorescence from anti-BrdU, AnnexinV/7AAD, and PI was measured by flow cytometry (BDPharMingen, San Jose, CA, USA) and analyzed by Flowjo (Tree Star, Ashland, OR, USA).
Western and Southern blotting
Lymphomas were lysed as previously described.23 Murine pre-B cells and fibroblasts were lysed in RIPA buffer (50 mM Tris, 150 mM sodium chloride, 1% TritonX-100, 0.1% SDS, 1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 40 μg/ml aprotinin, 10 mM β-gylcerophosphate, 1 mM sodium fluoride, 0.1 mM sodium vanadate). Equal amounts of protein were separated by SDS-PAGE. Antibodies specific for phosphorylated Erk1/2 (Cell Signaling, Danvers, MA, USA), total Erk1/2 (Cell Signaling), Ksr1 (BD Biosciences, Franklin Lakes, NJ, USA), phosphorylated Mek1/2 (Cell Signaling), total Mek1/2 (Cell Signaling), p19ARF (GeneTex, San Antonio, TX, USA), p53 (Ab-7, Calbiochem, La Jolla, CA, USA), Mdm2 (C-18, Santa Cruz Biotechnology, Santa Cruz, CA, USA), Myc (06-340, Upstate Biotechnology, New York, NY, USA), and β-actin (Sigma, St. Louis, MO, USA) were used to western blot. Horseradish peroxidase-linked secondary antibodies (GE Healthcare, Buckinghamshire, UK) and enhanced chemiluminescence (GE Healthcare) or Supersignal (Pierce, Rockford, IL, USA) were used to detect bound immunocomplexes. Densitometry for quantification of protein bands was performed using Image J software (National Institutes of Health, Bethesda, MD, USA). Band intensities of phospho-Mek and phospho-Erk are reported relative to band intensities of total Mek and Erk, respectively. Southern blots for Arf and p53 were preformed as previously described.23 Biallelic Arf deletions were determined by comparing the intensity of each band with the amount of DNA in the gel for that particular sample relative to the same in the controls (Arf wild-type and Arf deleted).
Abbreviations
- Arf:
-
alternative reading frame of the Ink4a locus
- BrdU:
-
bromodeoxyuridine
- EGF:
-
epidermal growth factor
- ERK:
-
extracellular signal-regulated kinase
- EtOH:
-
ethanol
- FACS:
-
fluorescence-activated cell sorting
- 4-OHT:
-
4-hydroxytamoxifen
- GFP:
-
green fluorescent protein
- IL-7:
-
interleukin-7
- IRES:
-
internal ribosome entry site
- Ksr1:
-
kinase suppressor of Ras 1
- Mapk:
-
mitogen-activated protein kinase
- Mdm2:
-
murine double minute 2
- MTT, (3-(4,5-Dimethylthiazol-2-yl)-2:
-
5-diphenyltetrazolium bromide
- MEF:
-
mouse embryonic fibroblast
- MEK:
-
mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- MSCV:
-
murine stem cell virus
- MycER:
-
Myc estrogen receptor fusion protein
- PI:
-
propidium iodide
- 7-AAD:
-
7-Amino-actinomycin D
References
Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985; 318: 533–538.
Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL . Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 1999; 13: 2658–2669.
Watanabe T, Ichikawa A, Saito H, Hotta T . Overexpression of the MDM2 oncogene in leukemia and lymphoma. Leuk Lymphoma 1996; 21: 391–397.
Young KH, Leroy K, Moller MB, Colleoni GW, Sanchez-Beato M, Kerbauy FR et al. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood 2008; 112: 3088–3098.
Moller MB, Ino Y, Gerdes AM, Skjodt K, Louis DN, Pedersen NT . Aberrations of the p53 pathway components p53, MDM2 and CDKN2A appear independent in diffuse large B cell lymphoma. Leukemia 1999; 13: 453–459.
McKay MM, Morrison DK . Integrating signals from RTKs to ERK/MAPK. Oncogene 2007; 26: 3113–3121.
Claperon A, Therrien M . KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene 2007; 26: 3143–3158.
Nguyen A, Burack WR, Stock JL, Kortum R, Chaika OV, Afkarian M et al. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol Cell Biol 2002; 22: 3035–3045.
Denouel-Galy A, Douville EM, Warne PH, Papin C, Laugier D, Calothy G et al. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr Biol 1998; 8: 46–55.
Kortum RL, Johnson HJ, Costanzo DL, Volle DJ, Razidlo GL, Fusello AM et al. The molecular scaffold kinase suppressor of Ras 1 is a modifier of RasV12-induced and replicative senescence. Mol Cell Biol 2006; 26: 2202–2214.
Lozano J, Xing R, Cai Z, Jensen HL, Trempus C, Mark W et al. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Res 2003; 63: 4232–4238.
Razidlo GL, Kortum RL, Haferbier JL, Lewis RE . Phosphorylation regulates KSR1 stability, ERK activation, and cell proliferation. J Biol Chem 2004; 279: 47808–47814.
Hirsch-Ginsberg C, LeMaistre AC, Kantarjian H, Talpaz M, Cork A, Freireich EJ et al. RAS mutations are rare events in Philadelphia chromosome-negative/bcr gene rearrangement-negative chronic myelogenous leukemia, but are prevalent in chronic myelomonocytic leukemia. Blood 1990; 76: 1214–1219.
Case M, Matheson E, Minto L, Hassan R, Harrison CJ, Bown N et al. Mutation of genes affecting the RAS pathway is common in childhood acute lymphoblastic leukemia. Cancer Res 2008; 68: 6803–6809.
Kiessling MK, Oberholzer PA, Mondal C, Karpova MB, Zipser MC, Lin WM et al. High-throughput mutation profiling of CTCL samples reveals KRAS and NRAS mutations sensitizing tumors toward inhibition of the RAS/RAF/MEK signaling cascade. Blood 2011; 117: 2433–2440.
Alexander WS, Bernard O, Cory S, Adams JM . Lymphomagenesis in E mu-myc transgenic mice can involve ras mutations. Oncogene 1989; 4: 575–581.
Schwartz RC, Stanton LW, Riley SC, Marcu KB, Witte ON . Synergism of v-myc and v-Ha-ras in the in vitro neoplastic progression of murine lymphoid cells. Mol Cell Biol 1986; 6: 3221–3231.
Langdon WY, Harris AW, Cory S . Acceleration of B-lymphoid tumorigenesis in E mu-myc transgenic mice by v-H-ras and v-raf but not v-abl. Oncogene Res 1989; 4: 253–258.
Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM et al. Modelling Myc inhibition as a cancer therapy. Nature 2008; 455: 679–683.
Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI . A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 1995; 23: 1686–1690.
Sears R, Leone G, DeGregori J, Nevins JR . Ras enhances Myc protein stability. Mol Cell 1999; 3: 169–179.
Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J 1998; 17: 5001–5014.
Alt JR, Greiner TC, Cleveland JL, Eischen CM . Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J 2003; 22: 1442–1450.
Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011; 471: 235–239.
Green MR, Aya-Bonilla C, Gandhi MK, Lea RA, Wellwood J, Wood P et al. Integrative genomic profiling reveals conserved genetic mechanisms for tumorigenesis in common entities of non-Hodgkin's lymphoma. Genes Chromosomes Cancer 2011; 50: 313–326.
Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL . Bax loss impairs myc-induced apoptosis and circumvents the selection of p53 mutations during myc-mediated lymphomagenesis. Mol Cell Biol 2001; 21: 7653–7662.
Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM . Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene 2008; 27: 1590–1598.
Strasser A, Harris AW, Bath ML, Cory S . Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 1990; 348: 331–333.
Xiao H, Zhang Q, Shen J, Bindokas V, Xing HR . Pharmacologic inactivation of kinase suppressor of Ras1 sensitizes epidermal growth factor receptor and oncogenic Ras-dependent tumors to ionizing radiation treatment. Mol Cancer Ther 2010; 9: 2724–2736.
Kim M, Yan Y, Kortum RL, Stoeger SM, Sgagias MK, Lee K et al. Expression of kinase suppressor of Ras1 enhances cisplatin-induced extracellular signal-regulated kinase activation and cisplatin sensitivity. Cancer Res 2005; 65: 3986–3992.
Eischen CM, Packham G, Nip J, Fee BE, Hiebert SW, Zambetti GP et al. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene 2001; 20: 6983–6993.
Eischen CM, Woo D, Roussel MF, Cleveland JL . Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol 2001; 21: 5063–5070.
Egle A, Harris AW, Bouillet P, Cory S . Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA 2004; 101: 6164–6169.
Felsher DW . Oncogene addiction versus oncogene amnesia: perhaps more than just a bad habit? Cancer Res 2008; 68: 3081–3086; discussion 3086.
Tran PT, Fan AC, Bendapudi PK, Koh S, Komatsubara K, Chen J et al. Combined inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PLoS One 2008; 3: e2125.
Xing HR, Cordon-Cardo C, Deng X, Tong W, Campodonico L, Fuks Z et al. Pharmacologic inactivation of kinase suppressor of ras-1 abrogates Ras-mediated pancreatic cancer. Nat Med 2003; 9: 1266–1268.
Llobet D, Eritja N, Domingo M, Bergada L, Mirantes C, Santacana M et al. KSR1 is overexpressed in endometrial carcinoma and regulates proliferation and TRAIL-induced apoptosis by modulating FLIP levels. Am J Pathol 2011; 178: 1529–1543.
Scholl FA, Dumesic PA, Barragan DI, Harada K, Charron J, Khavari PA . Selective role for Mek1 but not Mek2 in the induction of epidermal neoplasia. Cancer Res 2009; 69: 3772–3778.
Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell 2009; 16: 324–335.
Bhalla S, Evens AM, Dai B, Prachand S, Gordon LI, Gartenhaus RB . The novel anti-MEK small molecule AZD6244 induces BIM-dependent and AKT-independent apoptosis in diffuse large B-cell lymphoma. Blood 2011; 118: 1052–1061.
Acknowledgements
We would like to thank Maria Pia Arrate, Brandon Metge, and Rekha Kar for skilled technical assistance and members of the Eischen lab for helpful discussions. This work was supported by NCI grants R01CA148950 (CME), T32CA119925 (MWG), and the Cancer Center grant P30CA068485.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no conflict of interest.
Additional information
Edited by P Bouillet
Rights and permissions
About this article
Cite this article
Gramling, M., Eischen, C. Suppression of Ras/Mapk pathway signaling inhibits Myc-induced lymphomagenesis. Cell Death Differ 19, 1220–1227 (2012). https://doi.org/10.1038/cdd.2012.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2012.1
Keywords
This article is cited by
-
Targeting ST8SIA6-AS1 counteracts KRASG12C inhibitor resistance through abolishing the reciprocal activation of PLK1/c-Myc signaling
Experimental Hematology & Oncology (2023)
-
Super-enhancer-associated TMEM44-AS1 aggravated glioma progression by forming a positive feedback loop with Myc
Journal of Experimental & Clinical Cancer Research (2021)
-
Myc enhances B-cell receptor signaling in precancerous B cells and confers resistance to Btk inhibition
Oncogene (2017)
-
Scribble acts as an oncogene in Eμ-myc-driven lymphoma
Oncogene (2016)
-
praja2 regulates KSR1 stability and mitogenic signaling
Cell Death & Disease (2016)