Abstract
c-MET inhibitors are considered as a kind of novel drugs in non-small cell lung cancer (NSCLC) treatment. However, the results of different clinical studies involving c-MET inhibitors were not consistent. In this report, we performed Meta-analysis to investigate the beneficial and harmful effects of these drugs from 9 studies including 1611 patients in target drug groups and 1605 patients in control groups. As a result, patients in target drugs group had longer progression free survival (PFS) (HR 0.80, 95% CI 0.66–0.99, p = 0.04) but not overall survival (OS) than those in control group, especially in Asian (HR 0.57, 95% CI 0.42–0.76, p < 0.001), Non-squamous (HR 0.79, 95% CI 0.64–0.97, p = 0.03), Phase III (HR 0.66, 95% CI 0.50–0.86, p = 0.002), previous treated (HR 0.77, 95% CI 0.63–0.95, p = 0.01) and small molecular compounds subgroups (HR 0.62, 95% CI 0.50–0.78, p < 0.001). In addition, target drugs did not affect the objective response rate (ORR) but improved disease control rate (DCR) (RR 1.22, 95% CI 1.02–1.46, p = 0.03) of NSCLC patients. Our study first indicated that targeting c-MET therapies improved PFS and DCR in advanced or metastatic NSCLC patients, especially in previous treated Asian patients with adenocarcinoma.
Similar content being viewed by others
Introduction
As the leading cause of cancer-related death in the world, lung cancer is a major threat of health and heavy burden for family and society1,2. Traditionally, lung cancer is divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). The latter, accounting for nearly 80% of all lung cancer, can be further divided into squamous carcinoma, adenocarcinoma and large cell carcinoma by histology. However, this view should be renewed since the personalized medicine developed rapidly during the past decade3. It is of great importance to further classify NSCLC into specific subtypes with certain genetic markers, which is tightly related to therapeutic decision3. As the intrinsic trait of tumor cells, somatic mutation, chromosome rearrangement and copy number alterations existed in a large proportion of patients suffering from this disease4,5. Although the underlying mechanism of lung cancer has not been fully elucidated so far, it is widely accepted that some key genetic mutations in the airway epithelial cells play a pivotal role in the development of this malignancy.
There were many kinds of genomic aberrations observed in lung cancer patients, including epidermal growth factor receptor (EGFR) mutation and anaplastic lymphoma kinase (ALK) rearrangement, which are the most well known genetic alterations6,7. Comparatively, c-MET mutation is less common, and abnormal amplification of c-MET was found in about no more than 5% of NSCLC, mostly in adenocarcinoma8,9,10. Recent studies suggested that increased MET gene copy number or protein expression was conversely related to the prognosis of lung cancer, indicating a predictive value for this disease11,12. Subsequently, the drug inhibiting c-Met seems to be a new strategy for lung cancer management. In the past years, several kinds of drugs have been developed and applied into clinical trails, including tivantinib, crizotinib and onartuzumab etc. Nevertheless, the results of different clinical trails were not consistent13,14,15,16,17,18,19,20,21. For instance, the use of tivantinib prolonged the overall survival (OS) and progression-free survival (PFS) of patients with advanced lung cancer, while onartuzumab did not have an evident effect on PFS and OS during lung cancer therapy. The discrepancy might result from genetic background, different kinds of drugs and sample size. In order to determine the benefits and risks of the c-Met inhibitors, we conducted this meta-analysis to evaluate the efficacy and risk profiles of these drugs in lung cancer treatment.
Results
Characteristics of the included studies
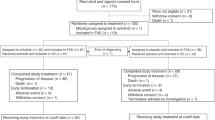
We identified 2270 relevant articles and abstracts, of which 73 studies were potentially suitable. 4 studies were eliminated due to lack of interest data, 24 were excluded because they were phase I or single-arm phase II trials, 26 were comments and reviews, 8 were retrospective studies and 2 studies with target drugs in both experimental and control arms. Thus, nine studies13,14,15,16,17,18,19,20,21, including 1611 patients from ten target drug groups and 1605 patients from ten control groups (the study by Wakelee et al. contained two experimental arms and two control arms), met the including criteria (Fig. 1). In addition, one article provided data of adverse events in several clinical trials about onartuzumab, which was not mentioned in other publications, so it was also included for our analysis22. Tivantinib and crizotinib were used in three and two studies, respectively, while onartuzumab was used as experimental drug in other four clinical trails. The study characteristics are listed in Table 1. All the patients were diagnosed by pathology and radiology. The quality of each study is listed in Table S1.
Overall survival
8 trails reported overall survival (OS) data, however, no difference of hazard ratio (HR) were observed between experimental group and control group (HR 0.99, 95% CI 0.89–1.10, p = 0.87) (Tables 2 and S3). Moreover, subgroup analysis showed that target drugs did not prolong overall survival of NSCLC patients in almost every subgroup, even in high MET expression group (Table S2).
Progression free survival
All studies presented progression free survival (PFS) results, and the patients in target drugs group had longer PFS than those in control group (HR 0.80, 95% CI 0.66–0.99, p = 0.04) (Tables 2 and S3). Further analysis indicated that lengthened PFS in Asian dominant (HR 0.57, 95% CI 0.42–0.76, p < 0.001), Non-squamous (HR 0.79, 95% CI 0.64–0.97, p = 0.03), Phase III (HR 0.66, 95% CI 0.50–0.86, p = 0.002), second or later line treatment subgroups (HR 0.77, 95% CI 0.63–0.95, p = 0.01). While small molecular compounds inhibiting c-MET significantly prolonged PFS (HR 0.62, 95% CI 0.50–0.78, p < 0.001), there were no difference of PFS between monoclonal antibody (onartuzumab) treatment group and control group(HR 1.05, 95% CI 0.91–1.21, p = 0.52). Unexpectedly, we also failed to observe the beneficial effect of target drugs in high MET expression group (HR 0.94, 95% CI 0.71–1.24, p = 0.52) (Table S2 and Fig. 2).
Subgroup analysis for progression-free survival (PFS) of NSCLC patients treated by c-MET inhibitors or placebo.
The studies were divided into two or more subgroups according to (a) ethnicity (Asian dominant vs. White dominant vs. Unknown), (b) histology (Squamous dominant vs. Non-squamous dominant), (c) phase of clinical trails (Phase II vs. Phase III), (d) prior treatment (without prior treatment vs. with prior treatment), and (e) drug types (small molecular compounds vs. monoclonal antibody). In addition, high MET expression subgroup (f) was also applied to analysis.
Objective response rate and disease control rate
The information of objective response rate and disease control rate were available in 8 and 5 studies, respectively (Table 2). Target drugs did not affect the ORR (RR 1.43, 95% CI 0.99–2.07, p = 0.06) but improved DCR (RR 1.22, 95% CI 1.02–1.46, p = 0.03) of NSCLC patients (Table S3 and Fig. 3).
Adverse events
Different kinds of adverse events were reported in each study, including anemia, neutropenia, rash, vomiting, diarrhea and so on. As a result, more patients with ILD (RR 4.47, 95% CI 1.15–17.45, p = 0.03, grade ≥ 3), edema (RR 3.23, 95% CI 2.24–4.64, p < 0.0001, all grades; RR 3.81, 95% CI 1.23–11.75, p = 0.02, grade ≥ 3) and respiratory infection (RR 2.24, 95% CI 1.63–3.07, p < 0.0001, all grades) was observed in target drug group. For other adverse events, there was no difference between experimental and control groups (Table 3).
Sensitivity analysis
Sensitivity analysis was performed to observe the effect of a single study on the overall results. No study was found to affect the pooled OS value significantly. However, the effect of prolonged PFS disappeared if several studies were omitted. In addition, three studies by Hirsch et al., Spigel et al. and Yoshioka et al. had effect on the result of ORR13,18,21, because an improved ORR value with statistical significance would be obtained while withdrawing any of these studies. As for DCR, all the trials dramatically influenced this index except the one by Yoshioka et al.21 (Figure S1).
Publication bias
Publication bias was tested using Begg’s and Egger’s tests. These tests did not show significant results in almost all comparisons (Table S3). The distribution of data points did not reveal evidence of obvious asymmetry (Fig. 4). These results indicated little publication bias.
Discussion
Lung cancer is the most common malignant tumor globally, and the five-year survival rate is less than 5%5. The ideal treatment of lung cancer is radical operation. However, a large proportion of patients lost the opportunity of surgery, because the tumors are in advanced or metastatic stage once they get the affirmative diagnosis23. For advanced lung cancer, drug treatment is inevitable. Platinum-based chemotherapy has been applied in lung cancer treatment for a long time24. Despite chemotherapy played effective role in some patients, its limit is becoming more and more obvious in widely clinical appliance, such as drug resistance, adverse effects etc.25. There is an urgent need for novel drugs.
Genetic alteration is responsible for the acquirement of malignant characteristics, such as proliferation without limitation, resistance to apoptosis, metastasis, and etc.26. On the other hand, various kinds of mutations also provide an attractive strategy for lung cancer treatment. Since the discovery of EGFR mutation and the effect of Gefitinib, the first generation of small molecular tyrosine kinase inhibitor targeting EGFR pathway, much efforts has been made to seek out other molecular targets and develop novel drugs in the past years27,28. Some investigators aimed at c-MET due to its abnormal amplification in some NSCLC patients, and some kinds of agents have been applied to clinical trials and obtained contradictory results7,9,29. However, no comprehensive assessment of c-MET-targeting therapy in NSCLC treatment has been carried out so far.
In our study, we first found that the agents targeting c-MET significantly prolonged PFS but not OS in advanced or metastatic NSCLC patients, which was in consistence with the majority of individual studies. We assumed that several factors might lead to this result. First, all the enrolled patients were in stage IIIB or IV, which meant a large proportion of subjects had metastatic tumors in other organs. For these patients, the overall survival was limited. The target drugs were not powerful enough to prolong OS evidently. Second, for any individual study, the follow-up period can not cover the whole survival of all enrolled patients. The limited observational period might not find the difference of OS between experiment and control arms. Objective response rate (ORR) and disease control rate (DCR) are other indexes for evaluating the efficacy of drug in clinical trails. Our present data suggested that the patients treated by target drugs have a much higher DCR but not ORR than those with placebo treatment. According to definition, the disease control status included stable disease (SD), other than complete response (CR) and partial response (PR). Our results indicated that it was beneficial for patients with c-MET inhibiting agents treatment to maintain steady state, which was in line with the results that target drugs improved PFS. However, the reason why target drugs failed to improve OS and ORR was not clear.
In light of the great heterogeneity among clinical trials which might be associated with race, prior treatment, histology and drug types, subgroup analysis was performed to distinguish the confound factors. These factors seemed not affect the OS because no improvement of this index in any subgroup. However, PFS was much longer in Asian dominant, Non-squamaous, Phase III, with prior treatment subgroups, which provided a hint for selecting proper subpopulations in future clinical application. Compared with monoclonal antibody, only small molecular compounds (tivantinib and crizotinib) exhibited beneficial therapeutic effects in our analysis. The most possible reason was the manner in which drugs exerted their effects. Tivantinib is the first non-ATP competitive small molecular compound that targets the inactive kinase domain of MET30. The unique structure of the agent leads to the high selective action with MET31. Crizotinib, initially designed as c-MET inhibitor, is usually applied in patients with ALK arrangement due to its multi-target actions32. Onartuzumab, however, is a recombinant, humanized, monovalent, monoclonal antibody that binds with MET, thus preventing this receptor from combining with its ligand, hepatocyte growth factor (HGF)33. The different pharmacological mechanisms might led to the significant variance of therapeutic effect. Otherwise, the selection of proper patients and treatment period should also be taken into account.
The existence of obvious inconsistency suggested the potential of influence on overall results. As we expected, sensitivity analysis showed that all the studies had minor effect on OS, but several studies did have significant effect on PFS, ORR and DCR. Five studies were demonstrated to have influence on PFS, which was a positive result presented in this meta-analysis14,15,16,17,21. Among these, four studies with relatively greater weight reported positive effect favoring c-MET inhibitor14,16,17,21. Subsequently, this effect of PFS disappeared once removing one of these studies. Similar results were also observed when we analyzed DCR. In converse, excluding three studies would make the overall effect of ORR, a negative result in our work, favor c-MET inhibitor13,18,21. In view of the discrepancy among these studies, the therapeutic effect of c-MET inhibitor was not robust enough, and more well-designed studies would contribute to getting more reliable results.
The most surprising finding in our analysis was that no positive results were observed in high met expression subgroup. In light that these agents were targeting c-MET signal pathway, the patients with high met expression were naturally considered as the favorable population. As a matter of fact, only Spigel et al. found a longer PFS in high met expression subgroup in a phase II study evaluating onartuzumab19. It was just this result that promoted the start of a large-scale phase III clinical trial in which all the enrolled patients were met positive18. Disappointingly, the expected results did not appear and the trial was stopped forever. In spite of this, we tried to find out the possible reasons. First, the evaluation methods on c-MET expression were not consistent among these studies. For the majority of included trials, immunohistochemistry (IHC) was applied to detect c-MET protein expression13,14,15,18,19,20,21, while cMET amplification was detected by fluorescent in situ hybridization (FISH) in another trial by Sequist et al.15. As a result, the stratification based on c-MET expression is not unified, which may affect overall results. Second, we noticed that not all the subjects in these trials have clear information on c-MET expression or amplification. Subsequently, evaluating the effect of target drug become more difficult since a large part of subjects is lack of c-MET information. Third, it is worth to note that c-MET might also interact with other oncogenic signal pathways due to the existence of multi-variations in an individual patient. For example, both Engelman and Bean found MET amplification led to resistance to EGFR targeting therapy in EGFR mutant patients with adenocarcinoma, indicating the potential relationship between c-MET and EGFR pathway34,35. Thus, it is more reasonable to compare the effect of target drugs between high met expression group and low met expression group under similar EGFR status. Nevertheless, the information of both c-MET and EGFR mutation in an individual subject was hardly available, which prevented us from exploring the real effect of target drugs. Anyway, this unusual phenomenon still needs further investigation.
Any drug not only brings patients benefits, but also side effects. Various kinds of adverse events were reported in each study adapted to our research. These drugs might have harmful effect on haemopoietic, circulatory, respiratory, digestive and nervous system. While most adverse events occurred equally in experimental and control arms, more cases of ILD, edema and respiratory infection were found in patients with target drug treatment. In addition, the adverse events seemed to be drug specific. For example, all the ILD were found in tivantinib and crizotinib group and most edema cases were reported in patients with onartuzumab, which might be associated with the intrinsic characteristics of different drugs. In addition, genetic polymorphisms of metabolic enzymes existed in Asian patients, so the administration doses of tivantinib were different in ATTENTIONAL study according to patients’ genetic background21. However, a fixed administration dose was applied to every subject in other trials without considering the individual variation. The inappropriate dose of drugs might also be a potential factor to adverse events.
Our study had some shortages. First, some clinical trials were still ongoing and only a part of data was published in academic meeting. The incomplete data restricted us from further research. Second, as the genetic background is an important factor to the development of lung cancer, it should be well controlled in clinical studies. However, a part of subjects in several studies have no information of genetic alteration. It might lead to the confusing results. Third, there were at least four kinds of small molecular compounds inhibiting c-MET, but other drugs were not put forward to RCT, except tivantinib and crizotinib, which prevented us from carrying out an exhaustive assessment for all c-MET inhibitors. Moreover, while most patients recruited to clinical study were Asian and Whites, almost no data on Africans was available. The incomprehensive data might affect the reliability of this study.
As a summary, our study first indicated that targeting c-MET therapies improved PFS and DCR but not OS and ORR in advanced or metastatic NSCLC patients, especially in previous treated Asian patients with adenocarcinoma. Compared with tivantinib and crizotinib, onartuzumab did not display a beneficial effect for NSCLC patients. Despite some limitations, our study suggested that anti-c-MET pathway was a promising method for NSCLC treatment.
Materials and Methods
Search strategy
We performed a comprehensive search strategy in databases, including PubMed and Embase, to identify the researches about drugs targeting c-MET and lung cancer. The terms we used were as follows: “non-small cell lung cancer”, “NSCLC”, “c-met inhibitor”, “tivantinib”, “crizotinib”, “carbozantinib”, “foretinib”, “onartuzumab”. Additional studies were identified by a manual search from references of original studies or review articles on this topic. In order to ensure the integrity of the related studies, the relevant abstracts from academic meetings were also retrieved other than full text articles.
Study Selection
The criteria for the selection of studies were as follows: (1) The studies should be randomized control trials (RCTs) with control arm and at least one experimental arm. (2) The drugs targeting c-MET in the studies should be used only in experimental arm. (3) The studies should provide at least one data of interest, such as overall survival (OS), progression free survival (PFS), objective response rate (ORR), disease control rate (DCR) and adverse effects (AE) etc. The studies in which the target drugs were used in both arms or the control arm was absent were excluded from our analysis.
Data extraction
The data were independently extracted from all eligible publications by two authors according to the inclusion criteria listed above. Any disagreements were resolved by discussions with a third person. The information were extracted from the studies include the basic information of clinical trial (trial number, author, year and phase), the traits of enrolled patients (quantity, ethnicity, genetic background, histological types and prior treatment), the regimens in both arms, the clinical outcomes (OS, PFS), and the frequencies of ORR, DCR and AE in each arm.
Quality assessment
All the enrolled trials were evaluated the quality with the help of the Cochrane five risk of bias domains tool, which included sequence generation, allocation concealment, data collection blinded, incomplete outcome data analyzing and selective outcome reporting. Each item was judged to obtain an assessment of “Yes” (low risk of bias), “No” (high risk of bias) or “Uncertain” (without enough information).
Statistical Analysis
Hazard ratio (HR) and 95% confidence intervals (CIs) were used to assess the relative risk of OS and PFS between experimental arm and control arm in the studies. While analyzing ORR, DCR and AE in target drug groups and control groups, relative risk (RR) and 95% confidence intervals (CIs) were applied.
I2 statistic was used to quantify the degree of heterogeneity, with I2 <25%, 25–75% and >75% representing low, moderate and high degrees of inconsistency, respectively36,37. In the analysis of pooled data, we used two different models according to the traits of the included studies: If no heterogeneity was found, a fixed effect model was adopted to determine the effects of target drugs or a random effect model was applied. If heterogeneity existed across studies, a subgroup analysis was performed to seek out the source of heterogeneity. The studies were subdivided by ethnicity (Asian dominant vs. White dominant), histological types (Squamous vs. Non-Squamous), prior treatment (first-line vs. second or later line), clinical trial phase (Phase II vs. Phase III) and different drug types (small molecular compounds vs. monoclonal antibody). Moreover, the patients with high MET expression were listed as a single subgroup. Sensitivity analysis was also performed to check the stability of the overall effect.
We made use of a Begg’s funnel plot to examine the underlying publication bias and Egger’s weighted regression method to calculate a P value for bias38,39. If no publication bias exists, the funnel plot appears symmetrical.
All analyses were conducted with the use of Review Manager, V.5.2 (Revman, The Cochrane Collaboration, London, UK) or STATA software, V.12.0 (StataCorp LP, College Station, TX, USA).
Additional Information
How to cite this article: Ye, S. et al. The Efficacy and Risk Profile of c-Met inhibitors in Non-small Cell Lung Cancer: a Meta-analysis. Sci. Rep. 6, 35770; doi: 10.1038/srep35770 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Dela Cruz, C. S., Tanoue, L. T. & Matthay, R. A. Lung cancer: epidemiology, etiology, and prevention. Clinics in chest medicine 32, 605–644 (2011).
Torre, L. A. et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians 65, 87–108 (2015).
Travis, W. D. et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer 10, 1243–1260 (2015).
Khanal, N. & Ganti, A. K. Emerging targeted therapies in non-small cell lung cancer. Expert review of anticancer therapy 16, 177–187 (2016).
Minuti, G., D’Incecco, A., Landi, L. & Cappuzzo, F. Protein kinase inhibitors to treat non-small-cell lung cancer. Expert opinion on pharmacotherapy 15, 1203–1213 (2014).
Rothschild, S. I. Targeted Therapies in Non-Small Cell Lung Cancer-Beyond EGFR and ALK. Cancers 7, 930–949 (2015).
Smyth, E. C., Sclafani, F. & Cunningham, D. Emerging molecular targets in oncology: clinical potential of MET/hepatocyte growth-factor inhibitors. OncoTargets and therapy 7, 1001–1014 (2014).
Garajova, I., Giovannetti, E., Biasco, G. & Peters, G. J. c-Met as a Target for Personalized Therapy. Translational oncogenomics 7, 13–31 (2015).
Gozdzik-Spychalska, J. et al. C-MET inhibitors in the treatment of lung cancer. Current treatment options in oncology 15, 670–682 (2014).
Padda, S., Neal, J. W. & Wakelee, H. A. MET inhibitors in combination with other therapies in non-small cell lung cancer. Translational lung cancer research 1, 238–253 (2012).
Dimou, A., Non, L., Chae, Y. K., Tester, W. J. & Syrigos, K. N. MET Gene Copy Number Predicts Worse Overall Survival in Patients with Non-Small Cell Lung Cancer (NSCLC); A Systematic Review and Meta-Analysis. PloS one 9 (2014).
Guo, B., Cen, H., Tan, X., Liu, W. & Ke, Q. Prognostic value of MET gene copy number and protein expression in patients with surgically resected non-small cell lung cancer: a meta-analysis of published literatures. PloS one 9, e99399 (2014).
Hirsch, F. R. et al. Efficacy and Safety Results From a Phase 2, Placebo-Controlled Study of Onartuzumab Plus First-Line Platinum-Doublet Chemotherapy in Advanced Squamous-Cell Non-Small Cell Lung Cancer (sq NSCLC). International Journal of Radiation Oncology*Biology*Physics 90, S37 (2014).
Scagliotti, G. et al. Phase III Multinational, Randomized, Double-Blind, Placebo-Controlled Study of Tivantinib (ARQ 197) Plus Erlotinib Versus Erlotinib Alone in Previously Treated Patients With Locally Advanced or Metastatic Nonsquamous Non-Small-Cell Lung Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 33, 2667–2674 (2015).
Sequist, L. V. et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 29, 3307–3315 (2011).
Shaw, A. T. et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. The New England journal of medicine 368, 2385–2394 (2013).
Solomon, B. J. et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. The New England journal of medicine 371, 2167–2177 (2014).
Spigel, D. R. et al. Onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIb or IV NSCLC: results from the pivotal phase III randomized, multicenter, placebo-controlled METLung (OAM4971g) global trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 32, abstr 8000 (2014).
Spigel, D. R. et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 31, 4105–4114 (2013).
Wakelee, H. A. et al. Efficacy and Safety of Onartuzumab in Combination With First-Line Bevacizumab- or Pemetrexed-Based Chemotherapy Regimens in Advanced Nonsquamous Non-Small Cell Lung Cancer (nsNSCLC): Results From a Phase 2, Placebo-Controlled Study (GO27821). International Journal of Radiation Oncology*Biology*Physics 90, S37–S38 (2014).
Yoshioka, H. et al. A randomized, double-blind, placebo-controlled, phase III trial of erlotinib with or without a c-Met inhibitor tivantinib (ARQ 197) in Asian patients with previously treated stage IIIB/IV nonsquamous nonsmall-cell lung cancer harboring wild-type epidermal growth factor receptor (ATTENTION study). Annals of oncology: official journal of the European Society for Medical Oncology/ESMO 26, 2066–2072 (2015).
Morley, R. et al. Safety of Onartuzumab in Patients with Solid Tumors: Experience to Date from the Onartuzumab Clinical Trial Program. PloS one 10, e0139679 (2015).
Goldstraw, P. et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer 2, 706–714 (2007).
Chang, A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung cancer 71, 3–10 (2011).
Stojanovska, V., Sakkal, S. & Nurgali, K. Platinum-based chemotherapy: gastrointestinal immunomodulation and enteric nervous system toxicity. Am J Physiol Gastrointest Liver Physiol 308, G223–G232 (2015).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Pirker, R. Novel drugs against non-small-cell lung cancer. Current opinion in oncology 26, 145–151 (2014).
Stinchcombe, T. E. Novel agents in development for advanced non-small cell lung cancer. Therapeutic advances in medical oncology 6, 240–253 (2014).
Sharma, N. & Adjei, A. A. In the clinic: ongoing clinical trials evaluating c-MET-inhibiting drugs. Therapeutic advances in medical oncology 3, S37–S50 (2011).
Adjei, A. A., Schwartz, B. & Garmey, E. Early clinical development of ARQ 197, a selective, non-ATP-competitive inhibitor targeting MET tyrosine kinase for the treatment of advanced cancers. The oncologist 16, 788–799 (2011).
Munshi, N. et al. ARQ 197, a novel and selective inhibitor of the human c-Met receptor tyrosine kinase with antitumor activity. Molecular cancer therapeutics 9, 1544–1553 (2010).
Ou, S. H. et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer 6, 942–946 (2011).
Merchant, M. et al. Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent. Proceedings of the National Academy of Sciences of the United States of America 110, E2987–E2996 (2013).
Bean, J. et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences of the United States of America 104, 20932–20937 (2007).
Engelman, J. A. et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007).
Higgins, J. P. & Thompson, S. G. Quantifying heterogeneity in a meta-analysis. Statistics in medicine 21, 1539–1558 (2002).
Higgins, J. P., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. Bmj 327, 557–560 (2003).
Begg, C. B. & Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994).
Egger, M., Davey Smith, G., Schneider, M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. Bmj 315, 629–634 (1997).
Acknowledgements
This work was supported by Zhejiang provincial Research Fund for Medical Sciences (No. 2012KYA011) and Natural Science Foundation of Zhejiang Province (No. LQ16H070003, No. LY17H160063).
Author information
Authors and Affiliations
Contributions
This study was designed by H.Z. The data were extracted by S.Y., J.L., K.H. and J.Y. performed statistical analysis and graph drafting, respectively. This manuscript was originally written and finally approved by all the authors.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ye, S., Li, J., Hao, K. et al. The Efficacy and Risk Profile of c-Met inhibitors in Non-small Cell Lung Cancer: a Meta-analysis. Sci Rep 6, 35770 (2016). https://doi.org/10.1038/srep35770
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep35770
This article is cited by
-
Differential responses of MET activations to MET kinase inhibitor and neutralizing antibody
Journal of Translational Medicine (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.