Abstract
The sex-determining gene SRY induces SOX9 expression in the testes of eutherian mammals via two pathways. SRY binds to testis-specific enhancer of Sox9 (TESCO) with SF1 to activate SOX9 transcription. SRY also up-regulates ER71 expression, and ER71 activates Sox9 transcription. After the initiation of testis differentiation, SOX9 enhances Amh expression by binding to its promoter with SF1. SOX8, SOX9 and SOX10, members of the SOXE gene family, also enhance the activities of the Amh promoter and TESCO. In this study, we investigated the regulation of these sexual differentiation genes in Tokudaia osimensis, which lacks a Y chromosome and the SRY gene. The activity of the AMH promoter was stimulated by SOXE genes and SF1. Mutant AMH promoters, with mutations in its SOX and SF1 binding sites, did not show significant activity by SOX9 and SF1. These results indicate that AMH expression was regulated by the binding of SOX9 and SF1. By contrast, SOXE genes could not enhance TESCO activity. These results indicate that TESCO enhancer activity was lost in this species. Furthermore, the activity of the SOX9 promoter was enhanced by ER71, indicating that ER71 may play an important role in the testis-specific expression of SOX9.
Similar content being viewed by others
Introduction
The master sex-determining gene SRY (sex-determining region Y) located on the Y chromosome is present in most eutherian mammals1,2. SRY initiates the transcription of SOX9 (SRY-box 9) in the genital ridge of the XY embryo, and an up-regulation of SOX9 expression gives rise to the Sertoli cells, resulting in testis development3. Sox9/SOX9 is necessary and sufficient for male sex determination in the mouse and human. In the mouse, SRY activates the testis-specific expression of Sox9 via two pathways. In the first mechanism, SRY binds to the enhancer TESCO (TES [testis-specific enhancer of Sox9] COre), which is located 13 kb upstream of Sox9 together with SF1 (also known as nuclear receptor subfamily 5, group A, member1, NR5A1), to induce Sox9 expression4. The TESCO sequence contains several SRY binding sites (BSs) and SF1 BSs that are highly conserved between the mouse, rat, dog, and human4. Sry expression is restricted to 10.5 and 12.5 days post-coitum (dpc) in the mouse5,6,7. Thereafter, SOX9 binds to SRY BSs in TESCO for its self-regulation4. In the second mechanism, SRY regulates Sox9 expression via Er71 (ETS related 71; also known as ETS variant 2, ETV2)8. SRY binds to the promoter region of Er71 with the transcriptional factor SP1 activates Er71 expression in the testes. ER71 subsequently regulates Sox9 expression by binding to the Sox9 proximal promoter. After the Sry expression, SOX9 binds to the Er71 promoter to control the expression of Er71. Thus, transcription of Er71 and Sox9 are co-regulated each other in the mouse9.
SOX9 directly regulates the expression of AMH (anti-Müllerian hormone; also known as Müllerian inhibitory substance, MIS). After the initiation of testes differentiation, AMH expression is induced in the Sertoli cells of eutherian mammals10,11,12. A previous study reported that approximately 370 bp of the Amh 5′ flanking region was essential for its expression from 12.5 dpc until an early postnatal stage in the male mouse13. This region, defined as the Amh proximal promoter, contains one SOX BS, two SF1 BSs, one GATA4 BS, and one WT1 BS14,15,16,17,18,19,20,21. The SOX BS is the most important region for Amh expression22. Furthermore, these BSs within the AMH promoter are conserved in several eutherian mammals and marsupials such as the wallaby, suggesting therians (eutherians and marsupials) share a common AMH regulatory mechanism23,24.
Other SOX genes might also have important functions in testicular differentiation. The SOX gene family consists of 20 members. They contain a HMG (high-mobility-group) domain that binds DNA25, and SOX genes are categorized into ten subgroups26. Among these, SOX8 (SRY-box 8), SOX9, and SOX10 (SRY-box 10) belong to the SOXE group. All SOXE genes are expressed during mammalian testis development25,27,28, and the structure of these proteins is highly conserved. In vitro studies demonstrate that SOX8 and SOX10 can stimulate the activities of the AMH promoter and TESCO such as SOX927,29. In addition, SOX8 regulates Sertoli cell function in the adult male mouse30. Overexpression of Sox10 results in female-to-male sex reversal in the XX mouse, and its duplication on human chromosome 22q13 causes 46, XX testicular disorders of sex development (DSD)31,32,33,34. These reports suggest that SOX8 and SOX10 might compensate for SOX9 function in male differentiation.
In this study, we investigated the mechanism of sexual differentiation in an SRY-absent mammal, the Amami spiny rat (Tokudaia osimensis). The sex chromosome constitution of this species is XO/XO, which is caused by the absence of the Y chromosome35,36,37,38. Furthermore, this species lacks the SRY gene39,40,41, suggesting that T. osimensis has a unique sex-determining mechanism42. Although SOX9 is important for the sexual differentiation of this species, the enhancer activity of T. osimensis TESCO is not promoted by SOX9 and SF143. We report that AMH expression is regulated by SOX9 and SF1 in T. osimensis, and that ER71 regulates SOX9 transcription, similar to that observed in the mouse. However, SOX8 and SOX10 failed to activate T. osimensis TESCO. Our results indicate that the mechanism of sexual differentiation following ER71 to AMH expression is highly conserved in this mammal.
Results
Gene sequences and proximal promoter regions are conserved
The sizes of the open reading frames and the corresponding amino acids of AMH, SOX8, SOX10, and ER71 of T. osimensis are shown in Table 1. Their nucleotide and amino acid sequences were highly similar to those of the mouse and rat (Table 1). In particular, the functional domain of each gene was highly conserved between the mouse and rat. The TGF-β domain of AMH in the mouse and rat was 98.9% and 97.0% homologous, respectively; the HMG domain of SOX8 and SOX10 was completely identical; and the ETS domain of ER71 was 98.8% and 100% homologous, respectively. The sequences of the AMH proximal promoter (−357/+13) and the SOX9 promoter (−451/+13) were determined in T. osimensis (Fig. S1). All BSs (one SOX BS, two SF1 BSs, one GATA4 BS, and one WT1 BS) in the AMH proximal promoter were conserved in T. osimensis (Fig. S1a). Five ETS BSs (−308/−305, −292/−289, −215/−212, −170/−167, and −33/−30) were found in the SOX9 promoter (Fig. S1b).
FISH mapping of AMH, SOX8, and SOX10
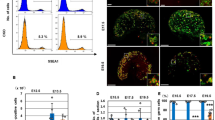
To determine the chromosomal locations of AMH, SOX8, and SOX10 in the T. osimensis genome, FISH mapping was performed. The T. osimensis BAC clones containing the open reading frames of AMH, SOX8, and SOX10, and the cDNA clone of each gene were used as probes. AMH, SOX8, and SOX10 were mapped to 8p13 (Fig. 1A,B), 3q12 (Fig. 1D,E), and 10q21 (Fig. 1G,H), respectively, by BAC FISH. To confirm that there were no duplicated copies of these genes in other loci, we performed also FISH mapping using cDNA clone of each gene. Similarly, each cDNA clone localized to the same chromosomal location (Fig. 1C,F,I). Each probe was found at a single locus in a pair of chromosomes.
Chromosomal localization of T. osimensis AMH, SOX8, and SOX10.
The AMH (A,B), SOX8 (D,E), and SOX9 (G,I) BAC clones, and the Amh (C), Sox8 (F), and Sox9 (I) cDNA clones were used as probes. Metaphase chromosomes were prepared from male T. osimensis. AMH, SOX8, and SOX10 were mapped to 8p13 (A–C), 3q12 (D–F), and 10q21 (G–I), respectively. The locations of specific gene signals were identical between BAC and cDNA clones. An arrowhead marks the hybridization signal. Propidium iodide-stained R- and Hoechst G-banding patterns are shown in (A,C,D,F,G,I) and (B,E,F,H) respectively. Scale bars represent 10 μm.
SOX9 induces AMH promoter transcriptional activity
The luciferase reporter construct (pGL3) containing the promoter region was co-transfected with the expression vector (pcDNA) into Cos7 cells. The generation of mouse Sox9 (mSOX9), T. osimensis SF1 (SF1), and T. osimensis SOX9 (TOS_SOX9) expression constructs were previously reported43. The luciferase vector containing the mouse Amh promoter (mAmh_pro) or the T. osimensis AMH proximal promoter (TOS_AMH_pro), and the different combinations of expression vectors were co-transfected into Cos7 cells. SF1 stimulated the activity of the positive control mAmh_pro by 4.5-fold compared to that observed for the empty vectors, whereas mSOX9 failed to up-regulate mAmh_pro activity (Fig. 2A). There was an approximately 7-fold increase in mAmh_pro activity after co-transfection with SF1 and mSOX9. These results agreed with a previous study14. Similarly, co-transfection with SF1 and TOS_SOX9 up-regulated TOS_AMH_pro activity (Fig. 2B).
SOX9 and SF1 activate the AMH promoter.
(A) Reporter gene activity of the mouse Amh promoter after co-transfection of SF1 and mSOX9 in Cos7 cells. A 7-fold increase in activity was observed after co-transfection with SF1 and mSOX9. (B) Reporter gene activity of the T. osimensis AMH promoter after transfection with SF1 or TOS_SOX9 in Cos7 cells. The AMH promoter was activated by SF1 and TOS_SOX9 in T. osimensis. The means ± SD from at least four independent experiments are shown.
Mutational analysis of the AMH promoter in T. osimensis
To determine whether SOX9 and SF1 bind to SOX BS and SF1 BS, respectively, to activate the AMH promoter, mutations were introduced in the proximal SF1 BS (R1; Regulatory mutation-1), the SOX BS (R2), and both SF1 BS and SOX BS in cis (R3)22,27 (Fig. S2). Mutations were introduced in TOS_AMH_pro by site-directed mutagenesis or splicing by overlap extension (SOE) PCR44. The mutated AMH promoters (R1, R2, and R3) were introduced into the pGL3 vector and co-transfected with SF1, TOS_SOX9, or both SF1 and TOS_SOX9 into Cos7 cells. The mutated TOS AMH promoters did not exhibit significant activity using different combinations of the expression constructs (Fig. 3).
SOX9 and SF1 bind to the T. osimensis AMH promoter.
The three AMH promoter mutants displayed low activity after co-transfection of different combinations of mouse and T. osimensis constructs. The fold-change in activity was calculated relative to the luciferase activity obtained from transfection with pGL3-empty constructs alone. R1, the mutation was introduced in the proximal SF1 BS within the AMH promoter of T. osimensis. R2, the mutation was introduced in the SOX BS. R3, the mutations were introduced in both SF1 BS and SOX BS in cis. The sequences of R1, R2, and R3 are shown in Fig. S3. The means ± SD from at least four independent experiments are shown.
SOX8 and SOX10 induce the transcriptional activities of the AMH promoter but not TESCO in T. osimensis
In the mouse, the Amh proximal promoter was activated by SF1 and SOX8, as well as by SF1 and SOX10 in vitro, similar to that observed for SF1 and SOX927,29. We investigated whether SOX8 and SOX10 could activate the AMH proximal promoter with SF1 in T. osimensis by a reporter gene assay. The open reading frames of mouse Sox8 and Sox10 (mSOX8 and mSOX10), and those of T. osimensis SOX8 and SOX10 (TOS_SOX8 and TOS_SOX10) were cloned into the pcDNA vector. The pGL3 vector containing mAmh_pro or TOS_AMH_pro, and the different combinations of expression vectors were transiently co-transfected into Cos7 cells. The mouse Amh promoter showed an approximately 7-fold increase in activity when co-transfected with both SF1 and mSOX8 or SF1 and mSOX10, similar to that observed for SF1 and mSOX9 (Fig. 4A). These results agreed with a previous study27,29. Similarly, TOS_AMH_pro was significantly activated by SF1 and TOS_SOX8, SF1 and TOS_SOX10, and SF1 and TOS_SOX9 (Fig. 4A).
SOX8 and SOX10, but not TESCO, activate the T. osimensis AMH promoter.
(A) The T. osimensis AMH promoter activity was enhanced by each combination of SOXE together with SF1, similar to that observed for the mouse. The fold-change in activity was calculated relative to the luciferase activity obtained from transfection with mAmh_pro alone. (B) T. osimensis TESCO displayed no enhancer activity for the different combinations of transfections. The fold-change in activity was calculated relative to the luciferase activity obtained from transfection with mTESCO alone. The means ± SD from at least four independent experiments are shown for both assays.
To determine whether SF1 and SOX8 or SF1 and SOX10 could stimulate the enhancer activity of T. osimensis TESCO, a reporter gene assay was performed. The pGL3 vector containing the promoter of mouse Sox9 and mouse TESCO (mTESCO) or T. osimensis TESCO (TOS_TESCO), which was prepared as previously described43, and the different combinations of expression vectors were transiently co-transfected into Cos7 cells. SF1 alone stimulated mouse TESCO activity by 3-fold compared with that of the empty expression vector, whereas mouse TESCO was not significantly activated by mSOX8, mSOX9, or mSOX10 alone (Fig. 4B). Mouse TESCO showed a greater than 4-fold increase in activity when co-transfected with SF1 and mSOX8, SF1 and mSOX9, or SF1 and mSOX10. These results agreed with a previous study29. Unlike mouse TESCO, TOS_TESCO did not exhibit significant activity using all combinations of the expression constructs (Fig. 4B). The SF1-mediated activities of TESCO were limited to approximately a 2-fold increase, and SF1 and TOS_SOX8, TOS_SOX9, and TOS_SOX10 failed to activate TOS_TESCO, resulting in a 2- to 2.5-fold increase in activity as previously reported43.
Expression of SOXE genes and ER71 in T. osimensis
The expression of Sox8/SOX8, Sox9/SOX9, Sox10/SOX10, and Er71/ER71 in several male and female mice and T. osimensis tissues was examined (Fig. S3). The expression patterns of the SOXE genes were mostly consistent with that of the mouse (Fig. S3A). However, testis-specific Er71/ER71 expression was observed in both the mouse and T. osimensis (Fig. S3B).
ER71 induces the transcriptional activity of the SOX9 promoter
A reporter gene assay was performed to determine whether ER71 can enhance the activity of the SOX9 proximal promoter in T. osimensis. The luciferase vectors containing the −453/+13 SOX9 proximal promoter of T. osimensis (TOS_SOX9_pro) or that of the mouse (mSOX9_pro), and the pcDNA containing the Er71 open reading frame of the mouse (mER71) or that of T. osimensis (TOS_ER71) were transiently co-transfected into Cos7 cells. For the positive control, the Sox9 promoter showed an approximately 2-fold increase in activity when co-transfected with ER71 expression constructs (Fig. 5A). TOS_SOX9_pro was also activated by ER71 in T. osimensis (Fig. 5B).
Discussion
The nucleotide and amino acid sequences of T. osimensis AMH, SOX8, SOX10, and ER71 were highly similar with those of the mouse and rat (Table 1). In addition, the functional domain of each gene in T. osimensis was highly homologous with that of rodent genes. Results from FISH mapping revealed that each gene existed as a single copy within the genome (Fig. S2), indicating evolutionary conservation in this species.
The AMH proximal promoter sequence was highly conserved in T. osimensis (Fig. S1A). The reporter gene assays showed each SOXE protein stimulated the activity of AMH promoter together with SF1 like mouse (Fig. 4A), indicating that SOXE genes might function in sexual differentiation in male spiny rats. To determine whether SOX9 and SF1 bind to SOX BS and SF1 BS, respectively, and activate the AMH promoter, we performed reporter gene assays using three AMH promoter mutants of SOX BS and proximal SF1 BS (R1, R2, and R3, Fig. S3). Promoter mutants significantly reduced the luciferase activity (Fig. 3), revealing that binding of SOX9 and SF1 to BS is essential for the regulation of AMH expression. These results confirmed that the regulation of AMH by SOXE genes such as SOX9, which is especially important, was conserved in T. osimensis.
By contrast, TESCO enhancer activity was not stimulated by the SOXE genes and SF1 (Fig. 4B). This result was consistent with a previous study that demonstrated loss of TESCO enhancer activity in T. osimensis, T. tokunoshimensis, and T. muenninki43. The loss of enhancer activity was caused by nucleotide substitutions of SRY BS and SF1 BS within TESCO, leading us to conclude that SOX8 and SOX10 failed to activate TESCO due to substitutions. Indeed, SRY was lost in T. osimensis and TESCO displayed no enhancer activity, whereas SOX9 was expressed in the testes (Fig. S3). Our results support an idea that SOX9 expression in the testes must be regulated via another enhancer in T. osimensi43. In human, 516–584 kb upstream duplication and 607.1–639.6 kb upstream deletion of SOX9 cause XX DSD in the absence of SRY and XY DSD, respectively45. These discoveries implying the existence of other enhancers that work in concordance with a testis-specific enhancer such as TESCO and/or other regulatory elements for the gonad-specific expression pattern of SOX9.
Five ETS BSs were identified in the SOX9 proximal promoter of T. osimensis, and three out of five were species-specific (−215/−212, −170/−167, and −33/−30; Fig. S1B). Results from the reporter gene assay showed ER71 to enhance SOX9 promoter activity, illustrating the function of the SOX9 promoter was conserved in this species (Fig. 5B). In addition, ER71 was expressed in T. osimensis testes (Fig. S3). These results indicated that ER71 expression is regulated by SOX9, and that the downstream molecular pathway of ER71 is highly conserved in T. osimensis in the absence of SRY expression.
In the mouse, SP1 binds to the Sry promoter to activate Sry transcription46,47. SP1 is a zinc finger transcriptional factor ubiquitously expressed48,49. SRY enhances the Er71 expression by binding with SP1 to its promoter region10. In this study, ER71 expression was detected in the T. osimensis testis, suggesting that another ER71-regulated gene has superseded the function of SRY. There is a possibility that SP1 and other SOX genes such as SOX3, SOX8, and SOX10 may trigger ER71 expression in fetal gonads. However, additional studies are needed to clarify the regulation of ER71 expression and to identify the new sex-determining gene in T. osimensis.
On the basis of several reports and theoretical considerations, the evolution of sex-determining genes is believed to proceed from less to more complex50,51, suggesting that molecular regulation of downstream genes are more highly conserved between taxonomic groups. Our results, which showed that the regulations of SOX9 by ER71 and AMH by SOX9 were highly conserved in the SRY-absent species (Fig. 6), support this contention. In conclusion, we showed that the molecular cascades involved in male sexual differentiation are highly conserved in the SRY-absent species. These findings contribute to evolutionary studies of sex-determining and sex-differentiating genes in eutherian mammals.
Schematic model for sexual differentiation in the SRY-absent mammal, T. osimensis.
In the male Amami spiny rat (T. osimensis), a new sex-determining gene superseded SRY. This gene might activate SOX9 via another enhancer (not TESCO) and ER71 during sexual differentiation. This study showed that the downstream cascade of SOX9 was conserved in this species.
Materials and Methods
Animals
T. osimensis, an endangered species (The IUCN Red List of Threatened Species; http://www.iucnredlist.org/1/1/2016), has been protected by the Japanese government as such since 1972. With permission from the Agency for Cultural Affairs, the Ministry of the Environment in Japan, T. osimensis were captured in cage traps on Amami-Oshima Island. To obtain fibroblasts for cell culture experiments, the tips of their tails were cut with surgical scissors. Tissues were harvested from animals that died naturally or accidentally. Total RNA was obtained from two females and two males. All the animal experiments in this study were approved by the Institutional Animal Care and Use Committee of the National University Corporation Hokkaido University and performed in accordance with the Guidelines for the Care and Use of Laboratory Animals, Hokkaido University.
Isolation of BAC clones containing AMH, SOX8, and SOX10
A T. osimensis BAC library was previously constructed43. PCR primer pairs were designed (Table S1) and used to screen the BAC library using a two-step 3D PCR screening system as previously described43. The isolated BAC clones of AMH, SOX8, and SOX10 were defined as TOB1-73N22, TOB1-283L22, and TOB1-65I6.
Cloning and sequencing of each gene and promoter
Total RNA was extracted from mouse and T. osimensis tissues using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. The total RNA was reverse transcribed using SuperScript III (Invitorgen) and oligo(dT) primers. The synthesized cDNA and BAC clones were used as templates for coding sequence (CDS) amplification and promoter sequence amplification, respectively. We designed primer pairs to amplify the coding regions of AMH, SOX8, SOX10 and ER71 and the promoter regions of AMH and SOX9 by comparing mouse and rat DNA sequences. The primer sequences are shown in Table S1. The GenBank accession number of each gene is as follows: LC149849 for AMH CDS, LC149850 for SOX8 CDS, LC149851 for SOX10 CDS, LC149852 for ER71 CDS, LC149853 for the AMH promoter, and LC149854 for the SOX9 promoter.
Preparation of chromosomes for FISH mapping
The R-banded chromosomes and BAC FISH were prepared as previously described43. FISH using cDNA probes was performed as earlier described38.
Construction of plasmids for promoter analysis
The pcDNA3.1 (+) (Invitrogen) expression vector was used to prepare the plasmids. The entire open reading frame of each gene was cloned into the Hind III/BamH I restriction sites of the expression vector. The expression vectors inserted Sox9/SOX9 (mSOX9 and TOS_SOX9) and SF1 were previously constructed43. The amino acid sequence of mouse and T. osimensis SF1 was identical; therefore, we used T. osimensis SF1 expression vectors in all experiments. The AMH promoter (−357 to +13) and the SOX9 promoter (−451 to +13) were ligated into the Xho I/BamH I restriction sites of the pGL3-basic vector (Promega). To generate a mutant AMH promoter reporter construct, which would have mutations in the SOX or SF1 BS as previously described22, site-directed mutagenesis and SOE PCR were performed44. The sequence of each primer is shown in Table S1.
Reporter gene assays
COS7 cells were cultured in DMEM supplemented with 10% fetal bovine serum at 37 °C in an atmosphere of 5% CO2. COS7 cells were seeded at a density of 0.5 × 105 per well in a 24-well plate 24 h prior to transfection. Transfection was performed using 1.5 μl of Lipofectamine 3000 (Invitorogen). To measure the activity of TESCO, the reporter construct (550 ng of pGL3_mTESCO_SOX9pro, mTESCO, pGL3_TOSTESCO_SOX9pro, or TOSTESCO), different combinations of the expression vector (110 ng) or the pRL Renilla luciferase control reporter vector (30 μg) (Promega) were transfected according to the manufacturer’s instructions. The quantity of the expression vector was increased to 220 ng with the empty pcDNA3.1 vector. The activity of the AMH promoter was measured by using the reporter construct (400 ng of pGL3_mAMHpro, mMMHpro, pGL3_TOSAMHpro, TOSAMHpro, pGL3_TOSAMHpro_ SF1BSmutated [R1], pGL3_TOSAMHpro_SOXBSmutated [R2], or pGL3_TOSAMHpro_ SF1BS/SOXBSmutated [R3]), different combinations of the expression vector (20 or 40 ng), and pRL (20 ng). The quantity of the expression vector was increased to 60 ng with the empty pcDNA3.1 vector. To measure activity of SOX9 promoter, either 430 ng of reporter construct (pGL3_MSOX9pro or mSOX9pro, pGL3_TOSSOX9pro or TOSSOX9pro), several combinations of 43 ng of each expression vectors, and 20 ng of pRL. The total amount of expression vector was adjusted to 43 ng by empty pcDNA3.1. Forty-eight hours after transfection, the reporter activities were measured by Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. The reporter activity was normalized to Renilla luciferase activity as an internal control. Each experiment was carried out four independent times.
Additional Information
How to cite this article: Otake, T. and Kuroiwa, A. Molecular mechanism of male differentiation is conserved in the SRY-absent mammal, Tokudaia osimensis. Sci. Rep. 6, 32874; doi: 10.1038/srep32874 (2016).
References
Gubbay, J. et al. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346, 2452250 (1990).
Sinclair, A. H. et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244 (1990).
Svingen, T. et al. Ex vivo magnetofection: a novel strategy for the study of gene function in mouse organogenesis. Dev. Dyn. 238, 956–964 (2009).
Sekido, R. & Lovell-Badge, R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453, 930–934 (2008).
Koopman, P., Münsterberg, A., Capel, B., Vivian, N. & Lovell-Badge, R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348, 450–452 (1990).
Hacker, A., Capel, B., Goodfellow, P. & Lovell-Badge, R. Expression of Sry, the mouse sex determining gene. Development 121, 1603–1614 (1995).
Jeske, Y. W., Mishina, Y., Cohen, D. R., Behringer, R. R. & Koopman, P. Analysis of the role of Amh and Fra1 in the Sry regulatory pathway. Mol. Reprod. Dev. 44, 153–158 (1996).
De Haro, L. & Janknecht, R. Cloning of the murine ER71 gene (Etsrp71) and initial characterization of its promoter. Genomics 85, 493–502 (2005).
DiTacchio, L. et al. Transcription factors ER71/ETV2 and SOX9 participate in a positive feedback loop in fetal and adult mouse testis. J. Biol. Chem. 287, 23657–23666 (2012).
Josso, N. & Picard, J. Y. Anti-Müllerian hormone. Physiol. Rev. 66, 1038–1090 (1986).
Josso, N. et al. Anti-müllerian hormone: the Jost factor. Recent. Prog. Horm. Res. 48, 1–59 (1993).
Vigier, B., Tran, D., du Buisson, F. D. M., Heyman, Y. & Josso, N. Use of monoclonal antibody techniques to study the ontogeny of bovine anti-Mullerian hormone. Reproduction 69, 207–214 (1983).
Beau, C. et al. In vivo analysis of the regulation of the anti-Müllerian hormone, as a marker of Sertoli cell differentiation during testicular development, reveals a multi-step process. Mol. Reprod. Dev. 59, 256–264 (2001).
De Santa Barbara, P. et al. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol. Cell Biol. 18, 6653–6665 (1998).
Giuili, G., Shen, W. & Ingraham, H. The nuclear receptor SF-1 mediates sexually dimorphic expression of Mullerian Inhibiting Substance, in vivo. Development 124, 1799–1807 (1997).
Arango, N. A., Lovell-Badge, R. & Behringer, R. R. Targeted Mutagenesis of the Endogenous Mouse Mis Gene Promoter. Cell 99, 409–419 (1999).
Nachtigal, M. W. et al. Wilms’ tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell 93, 445–454 (1998).
Johnson, P. A., Kent, T. R., Urick, M. E. & Giles, J. R. Expression and regulation of anti-mullerian hormone in an oviparous species, the hen. Biol. Reprod. 78, 13–19 (2008).
Monniaux, D. et al. Regulation of anti-Müllerian hormone production in domestic animals. Reprod. Fertil. Dev. 25, 1–16 (2012).
Viger, R., Mertineit, C., Trasler, J. & Nemer, M. Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Mullerian inhibiting substance promoter. Development 125, 2665–2675 (1998).
Shen, W. H., Moore, C. C., Ikeda, Y., Parker, K. L. & Ingraham, H. A. Nuclear receptor steroidogenic factor 1 regulates the müllerian inhibiting substance gene: a link to the sex determination cascade. Cell 77, 651–661 (1994).
Arango, N., Lovell-Badge, R. & Behringer, R. Targeted Mutagenesis of the Endogenous Mouse Mis Gene Promoter: In Vivo Definition of Genetic Pathways of Vertebrate Sexual Development. Cell 99, 409–419 (1999).
Lukas-Croisier, C. et al. Follicle-stimulating hormone increases testicular Anti-Mullerian hormone (AMH) production through sertoli cell proliferation and a nonclassical cyclic adenosine 5′-monophosphate-mediated activation of the AMH Gene. Mol. Endocrinol. 17, 550–561 (2003).
Pask, A. J. et al. Marsupial anti-Mullerian hormone gene structure, regulatory elements, and expression. Biol. Reprod. 70, 160–167 (2004).
Schepers, G. E., Teasdale, R. D. & Koopman, P. Twenty Pairs of Sox. Dev. Cell. 3, 167–170 (2002).
Bowles, J., Schepers, G. & Koopman, P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227, 239–255 (2000).
Schepers, G., Wilson, M., Wilhelm, D. & Koopman, P. SOX8 is expressed during testis differentiation in mice and synergizes with SF1 to activate the Amh promoter in vitro. J. Biol. Chem. 278, 28101–28108 (2003).
Cory, A. T., Boyer, A., Pilon, N., Lussier, J. G. & Silversides, D. W. Presumptive pre-Sertoli cells express genes involved in cell proliferation and cell signalling during a critical window in early testis differentiation. Mol. Reprod. Dev. 74, 1491–1504 (2007).
Polanco, J. C., Wilhelm, D., Davidson, T. L., Knight, D. & Koopman, P. Sox10 gain-of-function causes XX sex reversal in mice: Implications for human 22q-linked disorders of sex development. Hum. Mol. Genet. 19, 506–516 (2009).
O’Bryan, M. K. et al. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev. Biol. 316, 359–370 (2008).
Aleck, K. A., Argueso, L., Stone, J., Hackel, J. G. & Erickson, R. P. True hermaphroditism with partial duplication of chromosome 22 and without SRY. Am. J. Med. Genet. 85, 2–4 (1999).
Cantu, J. M. et al. Trisomy 22q12 leads to qter: “aneusomie de recombinaison” of a pericentric inversion. Ann. génétique. 24, 37–40 (1981).
Nicholl, R. M. et al. Trisomy 22 and intersex. Arch. Dis. Child. Fetal. Neonatal. Ed. 71, F57–F58 (1994).
Seeherunvong, T. et al. 46,XX sex reversal with partial duplication of chromosome arm 22q. Am. J. Med. Genet. A. 127A, 149–1451 (2004).
Honda, T., Suzuki, H. & Itoh, M. An unusual sex chromosome constitution found in the Amami spinous country-rat, Tokudaia osimensis. Japanese J. Genet. 52, 247–249 (1977).
Honda, T., Suzuki, H., Itoh, M. & Hayashi, K. Karyotypical differences of the Amami spinous country-rats, Tokudaia osimensis osimensis obtained from two neighbouring islands. Japanese J. Genet. 53, 297–299 (1978).
Kobayashi, T. et al. Centromere repositioning in the X chromosome of XO/XO mammals, Ryukyu spiny rat. Chromosome Res. 16, 587–593 (2008).
Kuroiwa, A., Ishiguchi, Y., Yamada, F., Shintaro, A. & Matsuda, Y. The process of a Y-loss event in an XO/XO mammal, the Ryukyu spiny rat. Chromosoma 119, 519–526 (2010).
Soullier, S., Hanni, C., Catzeflis, F., Berta, P. & Laudet, V. Male sex determination in the spiny rat Tokudaia osimensis (Rodentia: Muridae) is not Sry dependent. Mamm. Genome. 9, 590–592 (1998).
Sutou, S., Mitsui, Y. & Tsuchiya, K. Sex determination without the Y Chromosome in two Japanese rodents Tokudaia osimensis osimensis and Tokudaia osimensis spp. Mamm. Genome. 21, 17–21 (2001).
Murata, C., Yamada, F., Kawauchi, N., Matsuda, Y. & Kuroiwa, A. Multiple copies of SRY on the large Y chromosome of the Okinawa spiny rat, Tokudaia muenninki. Chromosome Res. 18, 623–634 (2010).
Kuroiwa, A. et al. Additional copies of CBX2 in the genomes of males of mammals lacking SRY, the Amami spiny rat (Tokudaia osimensis) and the Tokunoshima spiny rat (Tokudaia tokunoshimensis). Chromosome Res. 19, 635–644 (2011).
Kimura, R., Murata, C., Kuroki, Y. & Kuroiwa, A. Mutations in the Testis-Specific Enhancer of SOX9 in the SRY Independent Sex-Determining Mechanism in the Genus Tokudaia. PLoS One 9, e108779 (2014).
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989).
Kim, G. J. et al. Copy number variation of two separate regulatory regions upstream of SOX9 causes isolated 46,XY or 46,XX disorder of sex development. J. Med. Genet. 52, 240–247 (2015).
Assumpção, J. G. et al. A naturally occurring deletion in the SRY promoter region affecting the Sp1 binding site is associated with sex reversal. J. Endocrinol. Invest. 28, 651–656 (2014).
Desclozeaux, M. et al. Characterization of two Sp1 binding sites of the human sex determining SRY promoter. Biochim. Biophys. Acta. - Gene Struct. Expr. 1397, 247–252 (1998).
Bouwman, P. & Philipsen, S. Regulation of the activity of Sp1-related transcription factors. Mol. Cell Endocrinol. 195, 27–38 (2002).
Safe, S. & Abdelrahim, M. Sp transcription factor family and its role in cancer. Eur. J. Cancer. 41, 2438–2448 (2005).
Schartl, M. Sex chromosome evolution in non-mammalian vertebrates. Curr Opin Genet Dev. 14, 634–641 (2004).
Wilkins, A. S. Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays 17, 71–77 (1995).
Acknowledgements
The authors thank E. Chiba and R. Kimura for technical support, and C. Nishida for helpful suggestions in cell culture experiments.
Author information
Authors and Affiliations
Contributions
T.O. performed all the experiments, analyzed all the data, and drafted the manuscript. A.K. conceived and designed the study and participated in manuscript writing. All authors read and approved of the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Otake, T., Kuroiwa, A. Molecular mechanism of male differentiation is conserved in the SRY-absent mammal, Tokudaia osimensis. Sci Rep 6, 32874 (2016). https://doi.org/10.1038/srep32874
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep32874
This article is cited by
-
The impact of induced pluripotent stem cells in animal conservation
Veterinary Research Communications (2024)
-
Chromosomal-level assembly of Tokudaia osimensis, Tokudaia tokunoshimensis, and Tokudaia muenninki genomes
Scientific Data (2023)
-
Sexual dimorphism in brain transcriptomes of Amami spiny rats (Tokudaia osimensis): a rodent species where males lack the Y chromosome
BMC Genomics (2019)
-
Molecular cloning and mRNA expression pattern of \(\varvec{Sox}\) Sox 4 in Misgurnus anguillicaudatus
Journal of Genetics (2018)
-
Diverse and variable sex determination mechanisms in vertebrates
Science China Life Sciences (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.