Abstract
Embryonic stem cells (ESCs) are useful for the study of embryonic development. However, since research on naturally conceived human embryos is limited, non-human primate (NHP) embryos and NHP ESCs represent an excellent alternative to the corresponding human entities. Though, ESC lines derived from naturally conceived NHP embryos are still very rare. Here, we report the generation and characterization of four novel ESC lines derived from natural preimplantation embryos of the common marmoset monkey (Callithrix jacchus). For the first time we document derivation of NHP ESCs derived from morula stages. We show that quantitative chromosome-wise transcriptome analyses precisely reflect trisomies present in both morula-derived ESC lines. We also demonstrate that the female ESC lines exhibit different states of X-inactivation which is impressively reflected by the abundance of the lncRNA X inactive-specific transcript (XIST). The novel marmoset ESC lines will promote basic primate embryo and ESC studies as well as preclinical testing of ESC-based regenerative approaches in NHP.
Similar content being viewed by others
Introduction
Embryonic stem (ES) cells are generally derived from the inner cell mass of preimplantation embryos at the blastocyst stage1,2,3,4,5,6,7. If cultured under appropriate conditions, ES cells can proliferate indefinitely while maintaining a pluripotent state1. They can give rise to all somatic cell types deriving from the embryonic germ layers, i.e. ectoderm, mesoderm and endoderm, as well as germ cells.
Due to their in vitro differentiation capability human ES cells hold great promise for curative cell replacement approaches8, reviewed in refs 9, 10, 11, 12. However, before pluripotent stem cell-based therapies may become a routine clinical option to treat specific diseases, biosafety and efficacy issues have to be tested thoroughly. Due to the close phylogenetic relationship of non-human primates (NHP) to humans, NHP ES cells represent an excellent alternative to human ES cells with regard to basic and preclinical ES cell research reviewed in refs 13,14 and allow experiments not possible with human embryos15. Moreover, studies in NHP are often more relevant with regard to the human than studies in more distant species, which do not always reflect human physiology and anatomy in an adequate way reviewed in refs 16, 17, 18, 19. Preclinical testing of ES cell-based regenerative medicine would benefit from appropriate NHP models.
Rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques belong to the group of Old World monkeys and are the most frequently used NHP species in biomedical research20. The first non-human primate embryonic stem cell line was derived from rhesus macaque in 19953.
The common marmoset monkey (Callithrix jacchus) is a New World monkey endemic to Brazil. It has several significant practical and biological advantages compared to macaques. The marmoset monkey is small, easy to handle and free of zoonoses. These characteristics contribute to relatively low housing costs. Additionally, common marmosets have a significantly shorter generation time as well as a higher fecundity than macaques. Therefore, the common marmoset is the NHP model of choice for specific applications and purposes and nicely complements the macaque models. This is reflected by many studies performed in the fields of stem cell research21,22, reproductive biology23,24,25, developmental biology26,27, and neurobiology reviewed in refs 28,29. Embryo-derived pluripotent stem cells of the common marmoset were first published in 19962. After this initial publication of eight cell lines, the generation of three additional marmoset monkey ES cell lines was reported in 20056. The currently published set of marmoset ES cell lines was completed in 2009 when we published an additional marmoset ES cell line which was originally also generated by Erika Sasaki and colleagues7. Unfortunately, all marmoset ES cell lines generated by Thomson et al.2 are not available anymore (E-mail communication with WiCell, which served as distributor of these cells).
Here, we report the generation and characterization of four novel marmoset monkey ES cell lines derived from natural preimplantation embryos. Importantly, three of the ES cell lines were derived from morula stages, which has not been reported so far. We characterized the novel ES cell lines in comparison with the established marmoset ES cell line cjes0017.
Materials and Methods
Animals and embryo collection
Marmoset monkeys (Callithrix jacchus) were obtained from the self-sustaining breeding colony of the German Primate Center (Deutsches Primatenzentrum; DPZ) and kept as described previously30. All aspects of this study were approved by an external ethics committee (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, license numbers AZ 42502-04/066/06 and AZ 42502-04-14/1462). All methods were carried out in accordance with the approved guidelines.
Female common marmosets (n = 3) in the age range of 3.9 to 7.3 years were used as embryo donors. All females were kept pairwise with fertile males, and their reproductive cycles were basically monitored as described previously31. Collection of natural embryos was performed by flushing the uterus on day 4 to 6 after ovulation to obtain preimplantation embryos. The uterus was flushed either by invasive or minimal-invasive methods31. After invasive surgery the animals received antibiotic therapy (Duphamox 0.1 ml i.m.), and analgesic medication was given 30 minutes before surgery (Metacam 0.1 ml s.c.). The awakening period under a warming infrared lamp was monitored carefully and animals were retransferred to their cages only after full recovery. Invasive surgery was applied maximally six times per animal, and only to those animals with at least four unsuccessful previous minimal-invasive embryo collection attempts. Minimal-invasive embryo collection was performed under short term anesthesia with Diazepam (0.05 ml/animal) and Alfaxan (0.1 ml/100 g bodyweight).
Cell Culture
Mouse embryonic fibroblasts (MEFs)
Gamma-irradiated MEFs served as feeder cells and were obtained as described previously30.
Initial culture of recovered embryos
After recovery, preimplantation embryos were transferred to and kept in embryonic stem cell medium (ESM)30 [KO-DMEM (Gibco), 20% (v/v) KnockOut Serum Replacement (Gibco), 1% (v/v) Penicilline/Streptomycine (Gibco), 0,1% (v/v) Amphotericin B (Gibco), 1% (v/v) MEM Non-Essential Amino Acids Solution (Gibco), 2 mM GlutaMAX (Gibco), 50 μM 2-mercaptoethanol (Gibco)] until removal of the zona pellucida (ZP). All ex vivo processing of the embryos was done at 37 °C. The ZP was removed using pronase (2 mg/mL, Sigma #P8811) dissolved in KO-DMEM (Gibco). Embryos were first washed in a 100 μL drop of pronase solution, then transferred into another drop of pronase solution and kept there for 1–3 min until degradation of the ZP was observed. ZP-free embryos were immediately washed sequentially in four drops of ESM to remove the pronase and finally transferred onto MEFs in a 35 mm diameter well with ESM. Embryos were allowed to attach without any disturbances for three days before cultures were checked. If primary outgrowths were observed, the culture was continued for 2 to 3 weeks until further passaging. All pluripotent cells were cultured under hypoxic conditions (37 °C, 8% CO2, 5% O2) in ESM, and medium was changed every two to three days. Passaging of primary outgrowths and of resulting ES cells is described below.
Expansion and maintenance of embryonic stem cells
For further passaging of the primary outgrowths and ES cells, StemPro Accutase (Life Technologies, #A11105-01) was used. Briefly, cells of one well in a six-well plate were washed with PBS and incubated with 1 mL Accutase at 37 °C for 4 min. The cell suspension was transferred to 5 mL of pre-warmed ESM and the remaining feeder layer was washed with 3 mL ESM. Cells were pelleted (5 min, 200 × g, RT), resuspended in ESM and seeded onto fresh MEFs. Medium was changed every two to three days.
PCR for the detection of pluripotency associated genes
Oligonucleotides (Sigma) used for detection of mRNA coding for pluripotency associated genes are listed in Table S1. KOD Hot Start DNA Polymerase from Novagen was used according to manufacturer’s instructions.
Immunofluorescence
Immunofluorescence stainings were performed as described previously30. Antibodies and their dilutions are listed in Table S2.
AP live stain
For detection of Alkaline Phosphatase (AP), Alkaline Phosphatase Live Stain (Life Technologies, #A14353) was used. Briefly, growth medium was removed and the culture was washed with pre-warmed DMEM/F-12 two times for 2–3 minutes. Then a 1X AP Live Stain working solution was applied directly on to the cell culture and incubated for 20–30 minutes. The AP Live Stain was removed and pre-warmed DMEM/F-12 was applied to the culture prior to the visualization of fluorescent-labeled colonies under fluorescent microscopy using a standard FITC filter. Images were captured immediately.
Teratoma formation and analysis
For teratoma formation, 1–2 × 105 mouse embryonic feeder cells were combined with 8–9 × 105 ES cells in a final volume of 70 μL PBS. 60–75 μL Matrigel (Corning, #354277) were added to this cell suspension and injected subcutaneously into the inguinal region of male immunodeficient SCID/beige mice. Teratomas were retrieved 10–17 weeks, in one case 24 weeks after injection. Teratomas were immediately fixed after recovery in Bouin’s solution. After paraffin embedding, they were sectioned at 5 μm. Sections were then Hematoxylin and Eosin stained or processed for immunohistochemistry as described previously30.
Karyotyping
Karyotyping was performed by the Cytogenetic Laboratory in the Department of Human Genetics at the Universitätsklinikum Hamburg-Eppendorf (Germany) according to standard procedures. Briefly, for each cell line chromosome preparation was done from two or three 35 mm wells with ES colonies. ES cells from the wells were pooled before analysis. Then the cells were arrested with 0.2 μg/ml colcemid for 3 h and dissociated with 0.25% trypsin EDTA. For hypotonic treatment, cells were subjected to 55 mM KCl and fixed with methanol/acetic acid (3:1). For each cell line, 15 metaphases from GTG banded chromosome spreads were analysed under a light microscope at a 1000× magnification and at least four metaphases were karyotyped using a cytogenetic image analysis system (CytoVysion; Leica Biosystems). Karyotyping was done according to the chromosome assigning of Neusser et al.32. In case of suspected mosaicism or clonal aberration, 30 metaphases were analysed.
Transcriptome analysis
Two independent biological replicates were analyzed for each cell line. For sequencing, the RNA-samples were prepared with the “TruSeq RNA Sample Prep Kit v2” according to the manufacturer’s protocol (Illumina). Single read (50 bp) sequencing was conducted using a HiSeq 2000 (Illumina). Sequences were aligned to the genome reference sequence of Callithrix jacchus (Ensembl genome assembly 3.2.1) using the STAR alignment software (version 2.3.0e)33 allowing for 2 mismatches within 50 bases. Subsequently, filtering of unique hits and counting was conducted with SAMtools (version 0.1.18)34 and HTSeq (version 0.6.1p1)35. Read counts were analyzed in the R/Bioconductor environment (version 3.2, www.bioconductor.org) using the DESeq2 package (version 1.8.1)36. Candidate genes were filtered to a minimum of 2-fold change and FDR-corrected p-value < 0.05. Gene annotation was performed using Callithrix jacchus entries from Ensembl v78 (www.ensembl.org) via the biomaRt package (version 2.24.0)37. GO enrichment analysis on candidate genes was conducted with the Goseq package (version 1.2)38 using standard parameters. The data discussed in this paper are generated in compliance with the MIAME guidelines and have been deposited in NCBI’s Gene Expression Omnibus, accessible through GEO Series accession number GSE70897.
Results
Establishment and culture of novel embryonic stem cell lines
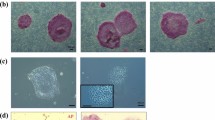
We established four embryonic stem cell lines, designated DPZcjESC1-4. The embryos were obtained from different donors (Table 1). Three of our novel lines were derived from morulae. DPZcjESC1 and 2 were obtained from early, non-compacted morulae, while DPZcjESC3 was obtained from a compacted morula. One line (DPZcjESC4) was derived from an expanded blastocyst stage embryo (Fig. 1A). In case of blastocysts, the embryos were plated onto the MEF layer with intact trophoblast. Immunosurgery to remove the trophoblast was not performed. All embryos formed clearly identifiable primary outgrowths (Fig. 1B). After passaging of the primary outgrowths and expansion on mouse embryonic feeder cells, all lines stably proliferated and exhibited the typical morphology of common marmoset pluripotent stem cells2,6,7,30,39. The cell lines formed tightly packed, flat colonies (Fig. 1C,D). Individual cells can be distinguished within a colony and the ES cells exhibit a high ratio of nucleus to cytoplasm. Prominent nucleoli are clearly visible (Fig. 1E). All cell lines tolerate enzymatic colony dissociation during passaging, which is done every six to 13 days. All cell lines tolerated cryopreservation, thawing and subsequent re-culturing.
(A) Embryos were obtained from adult common marmoset monkeys 4–6 days after ovulation. (B) Primary outgrowths on mouse embryonic feeder cells 13–15 days after plating of the embryo. (C) Colony overview, magnification 25-fold. (D) Single ES cell colony, magnification 200-fold. Individual passage number (P) is indicated in the upper left. (E) Magnification of framed area in D showing the prominent nucleoli. All bars = 100 µm.
Expression of pluripotency markers
To demonstrate the expression of characteristic pluripotency markers by the cell lines, we performed RT-PCR and immunofluorescence. On the mRNA level, we detected expression of SOX2, OCT4, KLF4, cMYC, NANOG, all being transcription factors, and LIN28, an RNA binding protein. All factors have been used for reprogramming of marmoset monkey somatic cells to a pluripotent state30,39. Messenger RNA coding for the pluripotency-associated transcription factor SALL4 was also detected. (Fig. 2A, Fig. S3). Expression of most of these marker genes was confirmed on the protein level by immunofluorescence (Fig. 2B). The ES cell lines also express the cell surface antigens TRA-1-81, TRA-1-60, SSEA-4 and SSEA-3 (Fig. 2B, Fig. S4) which identify pluripotent cells40. Moreover, expression of the transcription factor UTF141,42 as well as the Chromodomain Helicase DNA Binding Protein 1 (CHD1)43,44 was detected. Expression of the cell surface antigen SSEA-1, characteristic of mouse, but not primate pluripotent cells, was not detected (Fig. 2B, Fig. S4). All ES cell lines expressed alkaline phosphatase (Fig. 2C) and telomerase (Fig. 2D).
(A) PCR analysis of the generated ESC line DPZcjESC4. The ESC line expressed mRNA coding for well-established pluripotency-associated factors. Mouse embryonic feeder cells (MEFs) were used as negative control and the established embryonic stem cell line cjes0017 as positive control. (B) Immunofluorescence staining of ES cell colonies. Antibodies directed against pluripotency-associated epitopes confirmed expression of the surface molecules TRA-1-81, TRA-1-60, SSEA-4 and SSEA-3, the Chromodomain Helicase DNA Binding Protein 1 (CHD1), the RNA binding protein LIN28 and several transcription factors (NANOG, SOX2, OCT4, KLF4, SALL4, UTF1). As expected, SSEA-1 was not detected. (C) Alkaline Phosphatase (AP) live staining. Expression of AP was detected in all cell lines. (D) Normalized Telomerase expression (as obtained from the DeSEQ software) in the four novel ES cell lines DPZcjESC1-4, the established ES cell line cjes001 and primary skin fibroblasts. Transcripts coding for the Telomerase enzyme were detected only in ES cells. Data was generated by deep sequencing and analyzed with the DESeq2 package36. All bars = 100 µm.
Differentiation assay demonstrates pluripotency
To investigate the in vivo differentiation potential, we injected the ES cells into immunodeficient SCID/beige mice. Fig. 3 shows histological analysis of tumor tissue developed from the cell line DPZcjESC4. Ectodermal differentiation was indicated by neural tissue (Fig. 3A). Endoderm was represented by primitive gut-like epithelium (Fig. 3B) and mesoderm was represented by osteogenic (Fig. 3C′,C″), chondrogenic (Fig. 3C′) and hematopoietic differentiation (Fig. 3C″). Ectodermal differentiation was confirmed by immunohistochemical detection of β-Tubulin 3 (β-TUB III, ectoderm; Fig. 3A″) and endodermal differentiation by detection of SOX9 (Fig. 3B″, which is a marker of endodermal stem and progenitor cells). Histological analyses of teratoma tissue developed from the cell lines DPZcjESC1-3 are shown in Supplemental Figure S5. Together, these data show the pluripotent differentiation potential of the ES cells.
HE staining and immunohistochemical analysis of teratoma tissue derived from the cell line DPZcjESC4. HE staining demonstrates the presence of nerves (A), endodermal epithelium (B) and of mesodermal derivatives (C). (A′) Cross sections of peripheral nerves. (A′′) Detection of the neuronal marker β-Tub III. (A′′′) Isotype control for (A′′). (B′) Cross sections of a cyst lined with endodermal gut-like epithelium. (B′′) Detection of SOX9, which is a marker of endodermal progenitor cells. (B′′′) Isotype control for (B′′). (C′) Cartilage (lower left area) and bone trabecula (upper right corner). (C′′) Bone trabecula (upper part) and bone marrow including megakaroycytes (lower part). All bars = 100 µm.
Gene expression levels in morula- and blastocyst-derived cell lines
There were five ES cell lines (four novel and the reference cell line published previously7) available for this study. Two cell lines were derived from blastocysts (DPZcjESC4 and cjes001) and three from morula stages (DPZcjESC1-3). We performed whole transcriptome analyses of the marmoset ES cells by deep sequencing and included primary marmoset monkey fibroblasts as control. We were interested in whether the embryonic origin of the ES cell lines was reflected by differential mRNA expression of selected pluripotency markers by the corresponding ES cell lines. Hence, we analyzed expression levels of OCT4, SOX2 and NANOG as core pluripotency factors and further included LIN28, KLF4, KLF5, UTF1, DPPA3 (Stella), and GDF3. Analyses of the transcriptome data revealed similar relative expression levels of all markers analyzed in the individual samples of both the morula and the blastocyst-derived cell lines (Fig. 4A). To investigate whether global differences exist between the cell lines of different embryonic origin we performed a principle component analysis (PCA) providing a general overview (Fig. 4B): All four novel ES cell lines closely clustered together, irrespective of their embryonic origin. While the cell transcriptomes of different passages of the established ES cell line cjes001 had a higher data variance regarding component 2, the scatter was very low regarding the more indicative component 1 (46.39% variance). However, all ES cell lines were clearly separated from the fibroblasts. Hierarchical clustering on expression profiles confirmed that the novel ES cell lines are very similar but that their transcriptomes differ from the transcriptomes of the established ES cell line cjes001 (Fig. 4C). In a previous study where we generated common marmoset iPS cells by non-viral means30, hierarchical clustering showed that our iPS cells clustered apart from the cjes001. Interestingly, combining the global transcriptome data of both studies (the present and30) revealed that both our ES cells and iPS cell line clustered together and are more distant from the fibroblasts than cjes001 is (Fig. 4C).
(A) Expression levels of selected pluripotency markers. Levels of mRNA for OCT4, SOX2, NANOG, LIN28, KLF4, KLF5, UTF1, DPPA3 (Stella), and GDF3 in all four generated ES cell lines (DPZcjESC1-4) and in marmoset skin fibroblasts were compared to the levels in the established embryonic stem cell line cjes001. (B) Principal Component Analysis (PCA) plot of the four generated ES cell lines DPZcjESC1-4, the established ES cell line cjes001, the marmoset iPS cell line DPZcj_iPSC1, and primary skin fibroblasts showing the clustering of samples in gene expression space. Numbers in parentheses represent the percentage of total data variance covered by a principal component. (C) Heat map and hierarchical clustering of the normalized transcriptome expression profile for the generated ES cell lines DPZcjESC1-4, the previously generated iPS cell line DPZcj_iPSC1, the established ES cell line cjes001 and marmoset monkey fibroblasts. Darker color (blue) indicates higher correlation of gene expression.
Karyotyping
To the best of our knowledge, there is no official agreement on the chromosome assignment and nomenclature for the common marmoset monkey being comparable to the International System for Human Cytogenetic Nomenclature (ISCN, latest version 2013) which is updated every five years45. There is one study by Sherlock et al. published in 1996 suggesting a nomenclature46. However, it is very difficult to distinguish the g-banded chromosomes in the corresponding publication making an unambiguous chromosome assignment problematic. Therefore we refer to the study by Neusser et al.32 which in turn is relying on the nomenclature proposed by Sherlock et al.46. However, it must be pointed out that we are not able to unambiguously bring in line the banded chromosomes 19 and 20 from Neusser et al.32 with the idiograms presented in Sherlock et al.46.
Irrespective of the ambiguity associated with the assignment of marmoset chromosomes 19 and 20, we subjected all four ES cell lines to karyotype analysis. While the cell lines DPZcjESC3 and DPZcjESC4 had a normal female (46,XX) and male (46,XY) karyotype respectively, DPZcjESC1 (male) displayed a trisomy for chromosomes 18 and 19 (48,XY, +18, +19) and DPZcjESC2 (female) a trisomy for chromosome 19 (47,XX, +19) (Fig. 5).
Karyotype analysis of the four generated lines DPZcjESC1-4. The male cell line DPZcjESC1 displayed a trisomy for chromosomes 18 and 19 (48, XY, +18, +19), the female cell line DPZcjESC2 displayed a trisomy for chromosome 19 (47, XX, +19) and the cell lines DPZcjESC3 (female) and DPZcjESC4 (male) showed a normal karyotype (46, XX and 46, XY). Trisomic chromosomes are boxed.
The transcriptomes of the generated ES cells reflect their genomic status
Irrespective of their karyotype, all generated ES cell lines were indistinguishable from each other in all assays performed so far. Therefore we were wondering whether the additional copies of two chromosomes in DPZcjESC1 and of one chromosome in DPZcjESC2 might be subjected to regulatory mechanisms which lead to at least partial inactivation of the respective chromosome. This phenomenon of dosage compensation is known for the X chromosome (see below). Therefore we compared the relative expression per chromosome (calculated from the sum of all reads from one particular chromosome divided by the sum of all reads from all chromosomes) between the five ES cell lines. We found that the additional chromosomes are not subject to chromosome-wide regulatory mechanisms in the sense of dosage compensation since trisomies are clearly reflected by the deep sequencing data. Indeed, the relative expression of DPZcjESC1 was approximately 50% higher for two chromosomes, identified as 19 and 20 (p ~8 × 10−12 (paired t-test), blue arrows in Fig. 6A) in the transcriptome analysis. Likewise, the relative expression of DPZcjESC2 was ~1.5-fold (p = 1.26 × 10−6, (paired t-test), orange arrow in Fig. 6A) for chromosome 20. No such difference was found for any other chromosome that was present in disomy. Remarkably, karyotype analysis revealed trisomies for chromosomes 18 and 19 in DPZcjESC1 (instead of 19 and 20 as transcriptome analysis did) and a trisomy for chromosome 19 in DPZcjESC2 (instead of 20 as transcriptome analysis did).
(A) Relative average expression per chromosome. The relative average expression per chromosome was calculated from the sum of all reads from one particular chromosome divided by the sum of all reads from all chromosomes for each of the five ES cell lines. Blue arrows indicate significantly elevated expression from chromosomes 19 and 20 for DPZcjESC1, the orange arrow indicates significantly elevated expression from chromosome 20 for DPZcjESC2 and the black arrow indicates elevated expression from the X chromosome for cjes001. (B) Average expression of X-linked genes. Average reads per gene located on the X chromosome per million reads in the samples from the generated ES cell lines DPZcjESC1-4 and the established ES cell line cjes001. With 16.5 reads per million for the established female cell line cjes001 the average expression was 1.7-fold as high as for the four generated ES cell lines with an average of 9.5 reads per million (p-value of the t-tests was 3.29*10−6). C) X-inactive specific transcript (XIST)-expression. Number of reads mapping to the XIST-coding region detected in the samples from the generated ES cell lines DPZcjESC1-4 and the established ES cell line cjes001. Less than 3 reads per million were detected in the two male ES cell lines DPZcjESC1 and 4 whereas both female lines (DPZcjESC2 and 3) expressed XIST robustly (945 and 851 reads per million). In contrast, in the established ES cell line cjes001 XIST was not abundant (less than 5 reads per million).
We conclude that (i) two of our four new ES cell lines show trisomies, (ii) there is no evident compensatory gene expression (down-) regulation due to the presence of an additional copy of a chromosome, (iii) the transcriptome of the ES cell lines reflects their karyotype, (iv) it needs to be determined which chromosomes are affected (by clarification of numbering of the marmoset monkey chromosomes).
Different conditions of X-inactivation in female ES cell lines
Interestingly, comparison of the relative expression per chromosome between the five ES cell lines did not only confirm the trisomies determined by karyotyping. It also revealed that the average expression from the X-chromosome in the established female cell line cjes001 was much higher than in the novel ES cell lines (Fig. 6A, black arrow). That prompted us to analyze this issue in more detail. Quantitative analysis of the transcriptome data showed that the average expression of each gene on the X-chromosome in the established female cell line cjes001 was increased 1.7-fold compared to the average expression in the four novel ES cell lines (16.5 reads per million for cjes001 vs. an average of 9.5 reads per million for DPZcjESC1-4, p = 3.29 × 10−6, paired t-test) (Fig. 6B). X-inactive specific transcript (XIST) is a large long non-coding RNA known to be an important effector in the process of X inactivation47. We were wondering whether the significantly different average expression of the genes on the X chromosome would be associated with differences in the expression levels of XIST. Indeed, analysis of the transcriptome data revealed an evident correlation between XIST-expression and transcription activity from the X chromosome. While the relative amount of detected transcripts from the X chromosome was comparable in all ES cell lines generated in this study (Fig. 6A for an overview and Fig. 6B in higher resolution), almost no XIST (<3 reads per million) was detected in the two male ES cell lines (DPZcjESC1 and 4) whereas both female lines (DPZcjESC2 and 3) expressed XIST at high levels (945 and 851 reads per million) (Fig. 6C). In sharp contrast, in the previously established female ES cell line cjes001 where quantitative analysis of the transcriptome data indicated a significantly elevated transcriptional activity from the X chromosome, XIST was also almost absent being in the same range as in the male ES cell lines (<5 reads per million) (Fig. 6C).
We conclude that X-inactivation can occur in female ES cells of the common marmoset monkey and that X-inactivation correlates with expression of XIST. However, X-inactivation seems not to be an essential inherent characteristic of female ES cells.
Common marmoset ES cells exhibit features of primed pluripotency
Our novel ESC lines exhibit morphological characteristics of primed pluripotency, as the colonies are flat (Fig. 1) and not dome-shaped48. Further substantiating a primed state, the cells lack SSEA-1 expression and have an inactivated X chromosome48. In order to obtain insights into the molecular state of pluripotency (naïve versus primed) of the novel ES cell lines, we analyzed the presence of selected marker transcripts including REX1 (ZFP42), NROB1, FGF4, FGF5, and T (Brachyury) (Fig. 7). Since the regulation of pluripotency in the mouse is distinct from the regulation in primates27,48,49,50, the set of marker genes characterizing mouse naïve pluripotency51 does not exactly apply to primate pluripotent stem cells50. While REX1 and FGF4 are expressed in primed human pluripotent stem cells50,52,53, FGF5 is not upregulated in primed pluripotency50. Only little is known about NROB1 (also known as DAX1) in human pluripotent stem cells. However, mouse ES cells with a stable knockdown of NROB1 were predisposed to differentiation but still maintained pluripotency54. T is a marker of differentiating rhesus monkey ESCs55,56. REX1 expression was robust in all samples tested, and the average expression was comparable between the novel ESC lines, the previously established ESC line cjes0017, our previously generated iPSC line30 and neonatal and adult marmoset skin fibroblasts. In contrast, NROB1 expression was hardly detectable in any of the cell lines except for DPZcjESC4. Moderate FGF4 expression was detected in all ES cell lines and the iPS cell line but not in the fibroblasts. In contrast to FGF4, FGF5 expression was not detected in any of the novel ES and iPS cell lines, while very weak FGF5 expression was detected in the established ESC line cjes001 and in neonatal skin fibroblasts. Expression of Brachyury (T) was detected at different levels in all cell types except for the marmoset fibroblasts. In summary, quantitative analysis of these marker transcripts indicates a state of primed pluripotency of the novel marmoset monkey ES cells under the conditions used in this study.
Levels of mRNA for REX1, NROB1, FGF4, as markers of naïve pluripotency and FGF5, and Brachyury (T) as genes associated with primed pluripotency in all four generated ES cell lines (DPZcjESC1-4), the previously generated iPS cell line DPZcj_iPSC1 and in marmoset skin fibroblasts were compared to the levels in the established embryonic stem cell line cjes001.
Discussion
We report the derivation of four novel embryonic stem cell lines from natural preimplantation embryos of the common marmoset monkey (Callithrix jacchus). Although ESCs are usually derived from the inner cell mass of a blastocyst, it was previously shown that it is also possible to derive ESCs from embryos of earlier stages in mouse and human57, and even clonal lines from dissociated, single mouse and human blastomeres could be obtained58. However, to the best of our knowledge, all published non-human primate ES cell lines so far were derived from the inner cell mass of a blastocyst2,3,5,6,7. Here, we report for the first time the derivation of three marmoset ES cell lines from natural morula stage preimplantation embryos. One additional cell line was derived from a blastocyst stage embryo. Prior to culturing, we did not remove the trophoblast by immunosurgery as it is usually done during ES cell line establishment2,3,5,6,7. Our cells keep their pluripotent state and proliferate in standard embryonic stem cell medium, i.e. without the addition of bFGF or other growth factors.
We could not detect any obvious differences between morula- and blastocyst-derived cell lines – neither in morphology or growth characteristics nor on the molecular level. RT-qPCR did not reveal any statistically significant differences in the expression levels of selected pluripotency genes (data not shown) and also whole transcriptome analyses by deep sequencing proved the absence of global differences between the cell lines of different embryonic origin. In-depth analyses of the transcriptome data revealed differentially expressed genes. However, to prove that potential differences are due to the embryonic origin and not to other parameters, more (euploid) cell lines are needed.
Two of the four novel ES cell lines generated in this study, one male and one female line, have a normal karyotype while karyotype analyses revealed trisomies in the male cell line DPZcjESC1 and in the female cell line DPZcjESC2. According to karyotype analysis, chromosomes 18 and 19 are affected in DPZcjESC1, while global transcriptome data generated by deep sequencing and subsequent transcript mapping indicated trisomies of chromosomes 19 and 20. Analogously, in DPZcjESC2 there are three copies of either chromosome 19 or 20, depending on the method of analysis. Since we were not able to unambiguously bring in line the banded chromosomes 19 and 20 from two different publications32,46 and since the use of transcriptional profiling is a well-established technique59 we conclude that the karyotypes of our cells are [48, XY, +19, +20] for DPZcjESC1 and [47, XX, +20] for DPZcjESC2 as revealed by transcriptome analysis. However, an international agreement on a system for common marmoset monkey cytogenetic nomenclature according to the System for Human Cytogenetic Nomenclature45 would be highly desirable to prevent future confusion regarding cytogenetics in the common marmoset monkey.
Irrespective of the karyotypic characteristics of the four lines (that are also reflected in the transcriptomes), we did not observe any obvious differences between the euploid and aneuploid cell lines. Therefore, it still remains to be investigated in detail whether all cell lines have the same characteristics. Moreover, despite the unresolved issue of cytogenetic nomenclature, it would be very interesting to determine which human chromosomes correspond to the marmoset trisomic chromosomes in the novel ES cell lines in order to find out which human trisomies may be (partially) represented by the DPZcjESC lines 1 and 2. However, ignoring the difficulties of chromosome assignment and nomenclature described above, according to Sherlock et al.46, the entire marmoset chromosomes 18 and 19 show homology with human chromosome 1. This is particularly important because there are only a few autosomal trisomies that are compatible with survival to birth in humans60,61,62,63. Previously, it was suggested that the vast majority of human preimplantation embryos resulting from in vitro fertilization (IVF) are a mosaic of euploid and aneuploid blastomeres64. Tremendous 90% of the analyzed three- and four-day-old human cleavage stage embryos, most of them at the eight cell stage, showed chromosomal abnormalities. It was shown that large-scale structural chromosomal imbalances occurred in 70% of all embryos tested64. It is discussed that these high rates of aneuploidy observed in IVF embryos might either be an artefact of in vitro manipulation or that early human embryos undergo a unique, transient phase of chromosomal instability64. Interestingly, karyotype analysis showed that both of our cell lines obtained from early, non-compacted morulae (DPZcjESC1 and 2) are aneuploid while the remaining two cell lines (DPZcjESC3, DPZcjESC4) derived from later preimplantation stages, are euploid. Importantly, all four lines were obtained from natural, non-manipulated preimplantation embryos of the common marmoset monkey presaging that aneuploidy observed in human IVF embryos may also occur in naturally conceived primate embryos. While it is impossible to study naturally conceived early human embryos and since rodent embryos are not always an appropriate model for human embryology65, the marmoset could be a useful primate model species to further study the clinically relevant issue of aneuploidy in primate preimplantation embryos.
Transcriptome analysis of the ES cell lines revealed an evident inverse correlation between XIST-expression and transcriptional activity of X chromosomal genes. This allows the conclusion that X-inactivation in genotypically female ES cells of the common marmoset monkey occurs in a XIST-dependent manner. However, X inactivation does not seem to be an essential inherent characteristic for pluripotency in vitro66. Our data suggest that marmoset ES cells, with regard to X-inactivation, in general rather resemble human than mouse ES cells. For undifferentiated mouse ES cells, both X chromosomes are supposed to be active while X-inactivation was reported to be a feature of mouse ES cell differentiation67,68. In contrast to the mouse, human ES cells, which were reported to be generally in a primed state (in contrast to the mouse ES cells) do not display a consistent pattern of X-inactivation but rather show a high degree of variation between cell lines and even between sub-lines of the same parental cell line reviewed in ref. 69.
Quantitative analysis of the expression of marker genes for naïve or primed pluripotency of the common marmoset monkey ES cells suggests that the novel NHP ES cells are in a primed state of pluripotency (Fig. 7). We attempted to further characterize the state and the plasticity of the cells by culturing both euploid ES cell lines on Matrigel- and Vitronectin-coated plates under primed conditions70 and under naïve conditions in 2i-LIF media71. Under MEF-free primed conditions, the marmoset ES cells survived maximally for three days and then died (data not shown). Under the 2i-LIF conditions the cells proliferated and kept their colony-like morphology. However, the expression levels of OCT4A and NANOG dramatically decreased within a few days to less than 2% compared with the levels of cells cultured on MEFs (data not shown). We conclude that culture protocols established for human pluripotent cell are not directly applicable to these marmoset ES cell lines and that adaptations of the protocols for the feeder-free culture of human cells are necessary to enable feeder-free marmoset monkey ES cell culture.
The four novel ES cell lines derived in this study extend the available repertoire of marmoset monkey ES cell lines. Since ES cells derived from natural embryos are considered as the gold standard for pluripotent stem cells, their availability is crucial to identify fundamental (epigenetic) differences between primate ES and induced pluripotent stem (iPS) cells. Furthermore, pluripotency of the novel ESC lines 3 and 4, although apparently in a primed state under culture conditions currently established, may be worth testing in chimera formation assays.
Hierarchical clustering showed that the novel ES cell lines’ transcriptomes are very similar, but that they – together with the transcriptomes of an iPS cell line we previously generated30 – separate from the transcriptome of the established ES cell line cjes001. A deeper analysis revealed nearly 5000 differentially expressed genes between these two groups (our ES and iPS cells vs. cjes001 and fibroblasts). Top-20 enriched gene ontology (GO) terms are shown in Fig. S6 (our ES and iPS cells vs. cjes001 and fibroblasts) and S7 (our ES cells vs. cjes001). These data show that, although the culture conditions of the ES cell lines were identical and the cell lines phenotypically indistinguishable, there are clear differences between different cell lines on the transcriptomic level. This emphasizes the substantial need for the availability of an extended set of marmoset monkey ES cell lines for functional and preclinical studies.
Finally, two of our novel cell lines are female and two are male allowing the investigation of possible sex-specific differences between the lines. Interestingly, except for one ES cell line, which unfortunately is not available any more2, all ES cell lines from the common marmoset monkey published so far were female2,6,7.
Conclusion
In the context of the increasing interest in use of the common marmoset as a non-human primate model species that closely resembles human physiology (reviewed in refs 28,29), the four novel marmoset ES cell lines derived in this study are of great value. We previously generated marmoset monkey iPS cells by non-viral means30 with the long-term goal to develop and test cell replacement therapies in preclinical settings. Particularly in our ageing societies, cell replacement therapies based on pluripotent stem cells may become a promising option for the treatment of age-associated degenerative diseases. The availability of embryonic and induced pluripotent stem cells, both generated from the same marmoset colony, will allow elaborate comparative translational studies elucidating the potential of each cell type to restore functions of degenerated cells or tissues.
In summary, we have generated four novel ES cell lines from the common marmoset monkey, which exhibit significant differences with regard to their transcriptomes and genomes. The availability of these novel ES cell lines will further promote biomedical research in this NHP species, which is gaining more and more interest in translational studies including cell replacement therapies21,72.
Additional Information
How to cite this article: Debowski, K. et al. The transcriptomes of novel marmoset monkey embryonic stem cell lines reflect distinct genomic features. Sci. Rep. 6, 29122; doi: 10.1038/srep29122 (2016).
References
Evans, M. J. & Kaufman, M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981).
Thomson, J. A. et al. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol Reprod 55, 254–259 (1996).
Thomson, J. A. et al. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci USA 92, 7844–7848 (1995).
Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Suemori, H. et al. Establishment of embryonic stem cell lines from cynomolgus monkey blastocysts produced by IVF or ICSI. Dev Dyn 222, 273–279 (2001).
Sasaki, E. et al. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus). Stem Cells 23, 1304–1313 (2005).
Mueller, T. et al. A novel embryonic stem cell line derived from the common marmoset monkey (Callithrix jacchus) exhibiting germ cell-like characteristics. Hum Reprod 24, 1359–1372 (2009).
Kim, J. H. et al. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson’s disease. Nature 418, 50–56 (2002).
Karikkineth, B. C. & Zimmermann, W. H. Myocardial tissue engineering and heart muscle repair. Curr Pharm Biotechnol 14, 4–11 (2013).
Lakowski, J. et al. Transplantation of Photoreceptor Precursors Isolated via a Cell Surface Biomarker Panel from Embryonic Stem Cell-Derived Self-Forming Retina. Stem Cells (2015).
Russ, H. A. et al. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro . EMBO J 34, 1759–1772 (2015).
Grealish, S. et al. Human ESC-derived dopamine neurons show similar preclinical efficacy and potency to fetal neurons when grafted in a rat model of Parkinson’s disease. Cell Stem Cell 15, 653–665 (2014).
Joers, V. L. & Emborg, M. E. Preclinical assessment of stem cell therapies for neurological diseases. ILAR J 51, 24–41 (2009).
Wolf, D. P., Kuo, H. C., Pau, K. Y. & Lester, L. Progress with nonhuman primate embryonic stem cells. Biol Reprod 71, 1766–1771 (2004).
Tachibana, M. et al. Generation of chimeric rhesus monkeys. Cell 148, 285–295 (2012).
Kwiecien, T. D., Sy, C. & Ding, Y. Rodent models of ischemic stroke lack translational relevance… are baboon models the answer? Neurol Res 36, 417–422 (2014).
Pennesi, M. E., Neuringer, M. & Courtney, R. J. Animal models of age related macular degeneration. Mol Aspects Med 33, 487–509 (2012).
Iderberg, H., Francardo, V. & Pioli, E. Y. Animal models of L-DOPA-induced dyskinesia: an update on the current options. Neuroscience 211, 13–27 (2012).
Phillips, K. A. et al. Why primate models matter. Am J Primatol 76, 801–827 (2014).
National Research Council (US) Institute for Laboratory Animal Research, I. C. o. N. P. Animal Research in a Global Environment: Meeting the Challenges, Proceedings of the November 2008 International Workshop. (National Academies Press, US, 2008).
Kobayashi, Y. et al. Pre-evaluated safe human iPSC-derived neural stem cells promote functional recovery after spinal cord injury in common marmoset without tumorigenicity. PLoS One 7, e52787 (2012).
Shiozawa, S. et al. Gene targeting and subsequent site-specific transgenesis at the beta-actin (ACTB) locus in common marmoset embryonic stem cells. Stem Cells Dev 20, 1587–1599 (2011).
Wistuba, J. et al. Experimental endocrine manipulation by contraceptive regimen in the male marmoset (Callithrix jacchus). Reproduction 145, 439–451 (2013).
Vogt, E. J., Meglicki, M., Hartung, K. I., Borsuk, E. & Behr, R. Importance of the pluripotency factor LIN28 in the mammalian nucleolus during early embryonic development. Development 139, 4514–4523 (2012).
Lin, Z. Y. et al. Molecular signatures to define spermatogenic cells in common marmoset (Callithrix jacchus). Reproduction 143, 597–609 (2012).
Aeckerle, N., Drummer, C., Debowski, K., Viebahn, C. & Behr, R. Primordial germ cell development in the marmoset monkey as revealed by pluripotency factor expression: suggestion of a novel model of embryonic germ cell translocation. Mol Hum Reprod 21, 66–80 (2015).
Boroviak, T. et al. Lineage-Specific Profiling Delineates the Emergence and Progression of Naive Pluripotency in Mammalian Embryogenesis. Dev Cell 35, 366–382 (2015).
Okano, H., Hikishima, K., Iriki, A. & Sasaki, E. The common marmoset as a novel animal model system for biomedical and neuroscience research applications. Semin Fetal Neonatal Med 17, 336–340 (2012).
Kishi, N., Sato, K., Sasaki, E. & Okano, H. Common marmoset as a new model animal for neuroscience research and genome editing technology. Dev Growth Differ 56, 53–62 (2014).
Debowski, K. et al. Non-viral generation of marmoset monkey iPS cells by a six-factor-in-one-vector approach. PLoS One 10, e0118424 (2015).
Hanazawa, K. et al. Minimally invasive transabdominal collection of preimplantation embryos from the common marmoset monkey (Callithrix jacchus). Theriogenology 78, 811–816 (2012).
Neusser, M., Stanyon, R., Bigoni, F., Wienberg, J. & Muller, S. Molecular cytotaxonomy of New World monkeys (Platyrrhini) - comparative analysis of five species by multi-color chromosome painting gives evidence for a classification of Callimico goeldii within the family of Callitrichidae. Cytogenet Cell Genet 94, 206–215 (2001).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010).
Durinck, S. et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11, R14 (2010).
Tomioka, I. et al. Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors, including Lin28. Genes Cells 15, 959–969 (2010).
Lanctot, P. M., Gage, F. H. & Varki, A. P. The glycans of stem cells. Curr Opin Chem Biol 11, 373–380 (2007).
Pfannkuche, K., Fatima, A., Gupta, M. K., Dieterich, R. & Hescheler, J. Initial colony morphology-based selection for iPS cells derived from adult fibroblasts is substantially improved by temporary UTF1-based selection. PLoS One 5, e9580 (2010).
Laskowski, A. I. & Knoepfler, P. S. Myc binds the pluripotency factor Utf1 through the basic-helix-loop-helix leucine zipper domain. Biochem Biophys Res Commun 435, 551–556 (2013).
Gaspar-Maia, A. et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863–868 (2009).
Guzman-Ayala, M. et al. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development 142, 118–127 (2015).
Shaffer, L. G., McGowan-Jordan, J. & Schmid, M. ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013) (Karger Medical and Scientific Publishers, 2013).
Sherlock, J. K., Griffin, D. K., Delhanty, J. D. & Parrington, J. M. Homologies between human and marmoset (Callithrix jacchus) chromosomes revealed by comparative chromosome painting. Genomics 33, 214–219 (1996).
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S. & Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996).
Davidson, K. C., Mason, E. A. & Pera, M. F. The pluripotent state in mouse and human. Development 142, 3090–3099 (2015).
Frankenberg, S. R. Different Species Choose Their Own Paths to Pluripotency. Dev Cell 35, 267–268 (2015).
Weinberger, L., Ayyash, M., Novershtern, N. & Hanna, J. H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 17, 155–169 (2016).
Nichols, J. & Smith, A. Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 (2009).
Chan, E. M. et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol 27, 1033–1037 (2009).
Vallier, L., Reynolds, D. & Pedersen, R. A. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol 275, 403–421 (2004).
Zhang, J. et al. Dax1 and Nanog act in parallel to stabilize mouse embryonic stem cells and induced pluripotency. Nat Commun 5, 5042 (2014).
Schwanke, K. et al. Generation and characterization of functional cardiomyocytes from rhesus monkey embryonic stem cells. Stem Cells 24, 1423–1432 (2006).
Behr, R., Heneweer, C., Viebahn, C., Denker, H. W. & Thie, M. Epithelial-mesenchymal transition in colonies of rhesus monkey embryonic stem cells: a model for processes involved in gastrulation. Stem Cells 23, 805–816 (2005).
Strelchenko, N., Verlinsky, O., Kukharenko, V. & Verlinsky, Y. Morula-derived human embryonic stem cells. Reprod Biomed Online 9, 623–629 (2004).
Klimanskaya, I., Chung, Y., Becker, S., Lu, S. J. & Lanza, R. Human embryonic stem cell lines derived from single blastomeres. Nature 444, 481–485 (2006).
Ben-David, U., Mayshar, Y. & Benvenisty, N. Virtual karyotyping of pluripotent stem cells on the basis of their global gene expression profiles. Nat Protoc 8, 989–997 (2013).
Witters, G. et al. Trisomy 13, 18, 21, Triploidy and Turner syndrome: the 5T’s. Look at the hands. Facts Views Vis Obgyn 3, 15–21 (2011).
Cantu, E. S., Eicher, D. J., Pai, G. S., Donahue, C. J. & Harley, R. A. Mosaic vs. nonmosaic trisomy 9: report of a liveborn infant evaluated by fluorescence in situ hybridization and review of the literature. Am J Med Genet 62, 330–335 (1996).
James, R. S. & Jacobs, P. A. Molecular studies of the aetiology of trisomy 8 in spontaneous abortions and the liveborn population. Hum Genet 97, 283–286 (1996).
Sundareshan, T. S., Naguib, K. K., al-Awadi, S. A., Redha, M. A. & Hamoud, M. S. Apparently nonmosaic trisomy 22: clinical report and review. Am J Med Genet 36, 7–10 (1990).
Vanneste, E. et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med 15, 577–583 (2009).
Fougerousse, F. et al. Human-mouse differences in the embryonic expression patterns of developmental control genes and disease genes. Hum Mol Genet 9, 165–173 (2000).
de Oliveira Georges, J. A. et al. Aberrant patterns of X chromosome inactivation in a new line of human embryonic stem cells established in physiological oxygen concentrations. Stem Cell Rev 10, 472–479 (2014).
Guyochin, A. et al. Live cell imaging of the nascent inactive X chromosome during the early differentiation process of naive ES cells towards epiblast stem cells. PLoS One 9, e116109 (2014).
Schulz, E. G. et al. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 14, 203–216 (2014).
Payer, B. & Lee, J. T. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42, 733–772 (2008).
Vallier, L., Alexander, M. & Pedersen, R. A. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci 118, 4495–4509 (2005).
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Iwai, H. et al. Allogeneic Neural Stem/Progenitor Cells Derived From Embryonic Stem Cells Promote Functional Recovery After Transplantation Into Injured Spinal Cord of Nonhuman Primates. Stem Cells Transl Med 4, 708–719 (2015).
Acknowledgements
We are grateful to Angelina Berenson, Anna Magerhans, Nicole Umland and Simone Lüert for support and expert technical assistance. We thank Ellen Wiese for administrative support. The main parts of the study were done by institutional support from the German Primate Center (Deutsches Primatenzentrum, DPZ). The DPZ is a Leibniz-Institute which receives financial support from the Bundesrepublik Deutschland and the Bundesländer. Teratoma formation assays performed by Ralf Dressel were supported by the Collaborative Research Centre SFB1002 (TP C05) funded by the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Contributions
K.D. and R.B. were responsible for conception and design of the study, they wrote the main manuscript text. K.D. was responsible for cell culture and carried out analysis of the cell lines except for the aspects indicated below. C.D. was responsible for the embryo collection. J.L. and M.C. generated and interpreted qPCR data. R.D. performed the teratoma assays. G.S.-R. and T.L. were responsible for generation and interpretation of global transcriptome data. S.F. did the karyotyping analyses. E.S. provided the cell line cjes001. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Katharina Debowski is employed by STEMCELL Technologies SARL from March 1st 2016 on. All other authors declare that they have no competing interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Debowski, K., Drummer, C., Lentes, J. et al. The transcriptomes of novel marmoset monkey embryonic stem cell lines reflect distinct genomic features. Sci Rep 6, 29122 (2016). https://doi.org/10.1038/srep29122
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep29122
This article is cited by
-
A piggyBac-based platform for genome editing and clonal rhesus macaque iPSC line derivation
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.