Abstract
Neuregulin 4 (NRG4), an epidermal growth factor-like signaling molecule, plays an important role in cell-to-cell communication during tissue development. Its function to regulate energy metabolism has recently been reported. This current study was designed to assess the preventive and therapeutic effects of NRG4 overexpression on high fat diet (HFD)-induced obesity. Using the hydrodynamic gene transfer method, we demonstrate that Nrg4 gene transfer in mice suppressed the development of diet-induced obesity, but did not affect pre-existing adiposity and body weight in obese mice. Nrg4 gene transfer curbed HFD-induced hepatic steatosis by inhibiting lipogenesis and PPARγ-mediated lipid storage. Concurrently, overexpression of NRG4 reduced chronic inflammation in both preventive and treatment studies, evidenced by lower mRNA levels of macrophage marker genes including F4/80, Cd68, Cd11b, Cd11c, and macrophage chemokine Mcp1, resulting in improved insulin sensitivity. Collectively, these results demonstrate that overexpression of the Nrg4 gene by hydrodynamic gene delivery prevents HFD-induced weight gain and fatty liver, alleviates obesity-induced chronic inflammation and insulin resistance, and supports the health benefits of NRG4 in managing obesity and obesity-associated metabolic disorders.
Similar content being viewed by others
Introduction
Neuregulin 4 (NRG4) is a member of the neuregulin family (NRG1–NRG4) that shares a common structure of epidermal growth factor (EGF)-like domains1. Similar to other neuregulin members, NRG4 activates type-1 growth factor receptors (ErbB3 and ErbB4 receptor) to initiate cell-to-cell signaling through tyrosine phosphorylation1,2. NRG4 was originally detected in the adult pancreas and muscle, and is considered the essential factor for tissue development1. Further studies demonstrate that NRG4 is present in human breast milk and developing intestine tissue, promoting epithelial cell survival and protecting against experimental necrotizing enterocolitis3,4. In addition, NRG4 has been found in prostate, breast and gastric cancers5,6,7.
More recent work by Rosell et al. showed that NRG4 is enriched in brown adipose tissue (BAT) and its expression is upregulated in cold-induced beige/brite cells8. In vitro studies demonstrated that NRG4-contained medium from differentiated brown adipocytes promotes neurite outgrowth8. Using NRG4-deficient mice, Wang et al. revealed that NRG4 is capable of attenuating hepatic lipogenic signaling and maintaining metabolic homeostasis9. The findings from these previous studies indicate that NRG4 may work as a novel adipokine with a possible role in maintaining energy and metabolic homeostasis.
In the current study, we used a gene transfer system to enhance NRG4 expression and investigated its potential to prevent and to provide therapeutic benefits to animals with diet-induced obesity and metabolic disorders. Our results show that an increase of NRG4 expression inhibits diet-induced chronic inflammation, improves insulin resistance, and prevents weight gain in a diet-induced animal model.
Results
NRG4 gene transfer prevents diet-induced weight gain
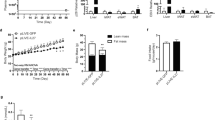
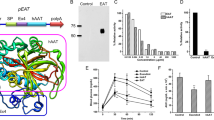
First, we confirmed the mRNA levels of Nrg4 in normal-weight and obese mice. Results in Fig. 1 show a reduction of the Nrg4 transcript level in the liver and epididymal white adipose tissue (eWAT) of the obese mice by ~40% and ~60%, respectively, compared to age-matched chow-fed mice. There was no difference in the Nrg4 transcript level in subcutaneous white adipose tissue (SubWAT) and BAT between normal-weight and obese mice. Results in Fig. 2a showed that hydrodynamic injections of pLIVE-NRG4 plasmids (on day 1 and day 28) were sufficient to protect mice from HFD-induced weight gain. After nine weeks of HFD feeding, pLIVE-NRG4-injected animals had an average body weight of 36 grams, 10 grams less than that of the control animals (~46 g) injected with control plasmid pLIVE-SEAP carrying the secreted embryonic alkaline phosphatase gene. The difference in body size was visually differentiable (Fig. 2c). Analysis of body composition revealed that the difference in body weight was primarily from fat mass (~49% reduction), not from lean mass (Fig. 2b). The average food intake between the two groups of animals was similar (Fig. 2d).
Eight-week-old C57BL/6 male mice were hydrodynamically injected via tail vein of 20 μg of pLIVE-NRG4 or pLIVE-SEAP control plasmid DNA (on days 1 and 28) and fed an HFD for 9 weeks. At the end of the experiment, total RNA was extracted from liver, EWAT, SubWAT, BAT, pancreatic tissues, and the relative mRNA levels of Nrg4 gene were determined by real-time or regular PCR. (a) Body weight-time curve; (b) Fat mass and lean mass at the end of 9-week feeding; (c) Representative images of mice at the end of the experiment; (d) Average food intake; (e) Detection of Nrg4 gene in the liver by regular PCR; and (f) Nrg4 mRNA level in different tissues determined by real time PCR. Each data point represents the mean ± SD of 5 animals. *P < 0.05, **P < 0.01 compared to control animals injected with pLIVE-SEAP plasmid DNA.
To test the effect of NRG4 increase on diet-induced obesity, we overexpress NRG4 by performing hydrodynamic gene delivery. Hydrodynamic-based gene transfer is an effective gene delivery method, which has been widely used for gene expression and functional analysis in whole animals10,11,12,13,14. Expression of NRG4 in animals receiving pLIVE-NRG4 plasmid transfer was confirmed by real-time PCR (RT-PCR) analysis and the results revealed that the mRNA level in the liver of the treated animals at the end of the experiment was approximately 97-fold higher than that of the control animals receiving two injections of pLIVE-SEAP control plasmids (Fig. 2f, Fig. S1). A high level of Nrg4 gene transfer was also confirmed by regular PCR analysis (Fig. 2e). In addition, an approximate 2–3-fold increase of Nrg4 expression was detected in the adipose tissues including BAT, eWAT, and SubWAT, but not in the pancreas (Fig. 2f). These results prove that NRG4 overexpression was responsible for blocking high fat diet-induced weight gain.
NRG4 suppresses chronic inflammation in eWAT and stimulates BAT thermogenesis
Consistent with the lower fat mass in pLIVE-NRG4-injected animals, H&E staining of adipocytes (Fig. 3a) and further quantitative analysis (Fig. 3b) revealed that Nrg4 gene transfer blocked the hypertrophy of adipocytes in eWAT and SubWAT by more than 50%, which was in line with the weight difference in the fat pads (Fig. 3c). Crown-like structures that are seen in the adipose tissue sections of the control animals are not detected in mice with the hydrodynamic delivery of pLIVE-NRG4 plasmids, indicating that NRG4 overexpression is capable of preventing diet-induced macrophage infiltration. Results from RT-PCR analysis (Fig. 3d) show that Nrg4 gene transfer suppressed HFD-induced expression of macrophage-specific marker genes including F4/80, Cd68, Cd11b and Cd11c by ~88%, ~87%, ~64% and ~83%, respectively. Nrg4 gene transfer also reduced monocyte chemotactic protein 1 (Mcp1) expression, a chemokine gene that regulates migration and infiltration of macrophages. Similarly, Nrg4 gene transfer enhanced M2 macrophage marker gene Cd163 (Supplemental Fig. S2). These results demonstrate the anti-inflammatory effect of NRG4 overexpression on diet-induced obesity. Nrg4 gene transfer also increased the expression of adiponectin (~3.2-fold) and adipose triglyceride lipase (Atgl, ~3.8-fold) genes.
Mice were sacrificed after 9 weeks of HFD feeding. Fat pads including EWAT, SubWAT and BAT were collected and weighed. Adipose tissues were fixed in 10% neutrally buffered formalin and H&E staining was performed. Total RNA was extracted from EWAT and BAT and relative mRNA levels of selected genes were determined by real-time PCR. (a) Representative images of H&E staining of EWAT, SubWAT and BAT (100×); (b) Average size of adipocytes in EWAT and SubWAT (calculated from measurements of 200 adipocytes from 5 separate slides); (c) Weight of different adipose pads; (d) Relative mRNA levels of selected macrophage marker genes including F4/80, Cd68, Cd11c, chemotactic factor gene Mcp1, Adiponectin and Atgl; (e) Relative mRNA levels of thermogenic genes including Ucp1, Ucp3, Pgc1α, Cidea and Dio2. *P < 0.05, **P < 0.01 compared to that of control animals injected with pLIVE-SEAP (n = 5).
Results in Fig. 3a (lowest panel) show fewer lipid droplets in BAT of pLIVE-NRG4-injected mice than those of the control animals, as determined by the density of vacuole-type structures. BAT is known as the thermogenic organ, facilitating utilization of extra glucose or lipids to generate heat15,16. To evaluate the effect of NRG4 overexpression on body temperature, we measured the rectal temperature of the animals with a digital thermometer. The results in Supplemental Fig. S1 showed that pLIVE-NRG4-injected mice had an increase of ~1.0 °C body temperature one day after the gene transfer and maintained this relatively higher body temperature for two weeks. Moreover, NRG4 overexpression also enhanced the expression of BAT thermogenic genes, including uncoupling protein Ucp1 (~ 2.2-fold), cell death-inducing DNA fragmentation factor-α-like effector A (Cidea, ~2.4-fold), and the type 2 deiodinase (Dio2, ~2.0-fold) (Fig. 3e), suggesting that Nrg4 gene transfer influences energy expenditure in BAT. Similar effects of Nrg4 gene transfer on browning marker genes were also seen in the inguinal WAT (Supplemental Fig. S4).
Nrg4 gene transfer prevents diet-induced hyperinsulinemia and insulin resistance
To assess the impact of Nrg4 gene transfer on insulin sensitivity and glucose homeostasis, glucose tolerance tests were performed and the results in Fig. 4a,b revealed a lower peak level and much higher clearance rate of intraperitoneally injected glucose in animals with Nrg4 gene transfer. pLIVE-NRG4-injected mice were more sensitive to insulin administration than the control mice (Fig. 4c). Nrg4 gene transfer dramatically suppressed an HFD-induced increase in the blood insulin level (7.3 μg/L vs. 1.6 μg/L, Fig. 4d). Compared to the control animals, pancreatic mRNA levels of Insulin1 and Insulin2 genes were markedly lower in mice with Nrg4 gene transfer (Fig. 4e). NRG4 overexpression also suppressed glucose 6-phosphate (G6p) gene expression, but not the phosphoenolpyruvate carboxykinase gene (Pepck) that is directly involved in glucogenesis (Fig. 4f). These results validate our hypothesis that NRG4 protects animals from obesity-associated insulin resistance.
(a) Time-dependent blood glucose level followed by IP injection of glucose (1.5 g/kg); (b) Area under the curve from glucose tolerance test (a); (c) Relative ratio of glucose concentration upon IP injection of insulin (0.75 U/kg); (d) Serum insulin levels at the end of the 9-week HFD feeding; (e) Relative mRNA levels of Insulin1 and Insulin2 in pancreas; (f) mRNA levels of G6p and Pepck gene involved in glucogenesis. Each data point represents the mean ± SD of 5 animals. *P < 0.05, **P < 0.01 compared to control animals.
Nrg4 gene transfer blocks hepatic lipogenesis and lipid storage
Obesity is associated with fatty liver disease17. Results in Fig. 5a show that Nrg4 gene transfer inhibits liver enlargement by 2.4-fold compared to that of the control animals (1.0 g vs. 2.4 g); a photograph of the liver discerned this outcome (Fig. 5b). H&E-stained liver sections showed a relatively normal liver structure in mice with Nrg4 gene transfer, compared to the extensive hepatocyte vacuolation in control animals (Fig. 5c). Oil Red O staining of liver sections also confirmed the difference (Fig. 5d). These results provide direct evidence in support that NRG4 overexpression is capable of blocking hepatic lipid accumulation.
At the end of the 9-week HFD feeding, mice were sacrificed and livers were collected and fixed for histochemistry. (a) Liver weight at the end of experiment; (b) Representative images of mouse livers; Liver sections were stained with H&E (c) or Oil Red O (d) (original magnifications 100×); (e) Relative mRNA levels of genes involved in hepatic lipid metabolism. Each data point represents the mean ± SD of 5 animals. *P < 0.05, **P < 0.01 compared to control animals.
We performed RT-PCR analyses to assess the effect of Nrg4 gene transfer on the expression of genes involved in hepatic lipid metabolism. Results in Fig. 5e show that NRG4 overexpression markedly suppressed the expression of lipogenic genes in the liver, including sterol regulatory element-binding protein 1c (Srebp1c, ~52%), acetyl-CoA carboxylase 1 (Acc1, ~37%), fatty acid synthase (Fas, ~38%), and stearoyl-CoA desaturase-1 (Scd1, ~89%). Nrg4 gene transfer also reduced the expression of peroxisome proliferator-activated receptor gamma (Pparγ1, ~77%), Pparγ2 (~99%), and their target genes, including Cd36 (~88%), fatty acid binding protein 4 (Fabp4, ~53%), and monoacylglycerol O-acyltransferase 1 (Mgat1, ~80%). In regard to cholesterol metabolism, NRG4 overexpression enhanced cholesterol 7 α-hydroxylase (Cyp7a1) gene expression by ~3-fold, but there was no change of the mRNA level for 3-hydroxy-3-methylglutaryl coenzyme A reductase (Hmgcr) and the ATP-binding cassette transporter (Abca1) genes (Fig. 5e). At the end of the 9-week experiment, Nrg4 gene transfer did not affect Pparα and its target gene, carnitine palmitoyl-transferase I (Cpt1). Collectively, these data demonstrated that prolonged Nrg4 gene expression alleviates obesity-associated fatty liver.
NRG4 overexpression improves insulin sensitivity in obese mice without changing body weight
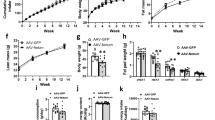
The preventive effect of NRG4 on HFD-induced obesity has been clearly demonstrated. Next, we assessed the effects of Nrg4 gene transfer on obese mice. Diet-induced obese mice (average body weight 56 g) were randomly divided into two groups and injected with pLIVE-NRG4 or pLIVE-SEAP plasmids using a hydrodynamics-based procedure. Results in Fig. 6a–c show that Nrg4 gene transfer failed to reduce body weight and did not change fat or lean mass. There was no difference in food intake between mice receiving pLIVE-NRG4 or pLIVE-SEAP plasmid DNA. However, NRG4-treated mice exhibited a much higher clearance rate of glucose in the glucose tolerance test (GTT) assay (Fig. 6d), which was better represented by the calculated area under the curve (AUC, Fig. 6e). Improvement of insulin sensitivity in obese mice was also seen in an insulin tolerance test. Taken together, these data demonstrate that Nrg4 gene transfer improves insulin sensitivity in obese mice.
C57BL/6 obese mice fed an HFD were hydrodynamically injected via tail vein with 20 μg of pLIVE-NRG4 or pLIVE-SEAP plasmid for 3 weeks. (a) Change of body weight; (b) Fat and lean mass measured 3 weeks after gene transfer; (c) Total food intake; (d) Time-dependent blood glucose level in glucose tolerance test (1.5 g/kg); (e) Area under the curve from glucose tolerance test; (f) Relative ratio of glucose concentration in insulin tolerance test (0.75 U/kg). *P < 0.05, **P < 0.01 compared to control animals (n = 5).
NRG4 overexpression reduced chronic inflammation in obese mice
H&E staining was performed and the results in Fig. 7a–c show no structural change of the liver, BAT, and SubWAT among the treated or control animals. Large lipid droplets in the liver and adipocytes were found in both groups of animals, indicating that lipid storage in these tissues was not affected. Gene expression analysis showed that Nrg4 gene transfer did not change the mRNA levels of Srebp1c and Acc1, but did reduce Fas and Scd1 mRNA levels by ~30% and ~42%, respectively (Fig. 7d). In addition, NRG4 overexpression reduced mRNA levels of Pparγ1 (~22%) and Mgat1 (~28%), but not that of Pparγ2 and Cd36 (Fig. 7e).
Results in Fig. 8a show significant macrophage infiltration into EWAT of the obese animals, as reflected by multiple crown-like structures in H&E-stained EWAT sections. However, the crown-like structures were less obvious in animals with Nrg4 gene transfer. RT-PCR was carried out to confirm the difference in the macrophage activation in the WAT of the treated and controlled animals. Results in Fig. 8b show a significant reduction in the mRNA levels of macrophage marker genes in mice with Nrg4 gene transfer, including F4/80, Cd68, Cd11b, and Cd11c in EWAT of obese mice. In addition, the mRNA level of inflammatory factor gene Mcp1 was also reduced by ~40%. A similar effect of Nrg4 gene transfer on gene expression was detected in the liver and BAT of mice with NRG4 overexpression (Fig. 8c,d). These results suggest that NRG4 overexpression is capable of inhibiting obesity-related chronic inflammation.
At the end of the 3-week. after gene transfer, obese animals were sacrificed. (a) Images of EWAT stained with H&E; (b–d) Relative mRNA levels of macrophage marker genes in EWAT, liver, and BAT including F4/80, Cd68, Cd11b, Cd11c and inflammatory factor Mcp1 and Tnfα. *P < 0.05, **P < 0.01 compared to control animals (n = 5).
Discussion
In this study, we systematically examined the effects of Nrg4 gene transfer on high fat diet-induced obesity and obesity-associated metabolic changes. The results show that Nrg4 gene transfer protects mice from diet-induced weight gain and adiposity (Figs 2 and 3), and inhibits fatty liver and insulin resistance (Figs 4 and 5). The beneficial effects are correlated with an inhibition of lipogenesis, lipid storage, and chronic inflammation (Figs 3 and 5). NRG4 overexpression in obese mice did not affect pre-existing fat and body weight, but reduced chronic inflammation and improved obesity-related insulin resistance (Figs 6, 7, 8).
Obesity increases the risk of type 2 diabetes, cardiovascular diseases, and certain cancers, which are among the leading causes of death worldwide18. Although a cure for obesity is important, treatments are costly, and success so far is limited due to a lack of safe and effective methods. Prevention that is capable of benefiting a larger majority of the population and halting obesity-related metabolic disorders is more desirable and effective. Weight control programs through dietary intervention and enhanced physical activities have been popular in the past, but the successes have been limited to a small percentage of people since persistence in restricting food intake and maintaining regular exercise can be difficult to accomplish19. Evidently, efforts to explore new methods and strategies are urgently needed in dealing with the obesity epidemic.
NRG4 was originally identified as a factor that plays a critical role in tissue development1. However, a high level of expression in BAT and cold-induced expression in WAT suggest that NRG4 may act as an adipokine with a distinct function8. Since BAT plays an essential role in regulating body temperature and thermogenesis15,16, as well as maintaining systemic lipid and glucose homeostasis20,21,22, an elevated level of NRG4 in BAT indicates that NRG4 may be involved in energy metabolism and maintenance of metabolic homeostasis. A lower mRNA level of Nrg4 (Fig. 1) in EWAT and in the liver of diet-induced obese mice appears to support such a prediction. In addition, a decrease in NRG4 levels was also observed in patients with non-alcoholic fatty liver disease (NAFLD)23.
NRG4 significantly inhibited lipogenesis by suppressing Srebp1c and its target genes, which is consistent with the previous results obtained from NRG4-deficient mice9. Elevating SREBP1c expression promotes lipid accumulation in the liver and fat tissue24,25, and HFD feeding induces SREBP1c-mediated lipogenesis in obese mice26,27. However, inhibition of lipogenesis exerts a positive effect on metabolic diseases28,29. Therefore, suppression of lipogenesis by NRG4 plays, at least in part, an important role in preventing diet-induced obesity. In addition, Nrg4 gene transfer inhibited expression of Pparγ and its target genes (Fig. 5e). PPARγ is a transcription factor regulating the expression of genes responsible for lipid storage and adipocyte differentiation30. Previous studies have shown that elevated expressions of Pparγ, Cd36, and Mgat1 genes are directly linked to hepatic steatosis31,32,33. Therefore, by targeting the hepatic PPARγ pathway, NRG4 protects against obesity-induced hepatic steatosis.
Several studies have demonstrated the interaction of NRG4 with inflammation. NRG4 levels in animals with ulcerative colitis (UC) and Crohn’s disease are reduced compared to un-inflamed controls. NRG4 treatment blocked inflammatory cytokine-induced apoptosis in colon epithelial cells and protected animals from experimental inflammatory bowel disease3,4. This conclusion agrees with our observation that NRG4 significantly reduced crown-like structures in EWAT and lowered expression of macrophage marker genes such as F4/80, Cd68, Cd11, and inflammatory factor Mcp1 (Figs 3 and 8). These results also align with our previous reports that suppressing HFD-induced inflammation by either antioxidant34 or macrophage elimination35 is sufficient to block HFD-induced obesity. However, results shown in Fig. 6a,b suggest that blockage of inflammation is not sufficient to reduce pre-existing body weight and fat mass in obese mice, indicating further strategy is needed to mobilize stored fat.
Reduction of inflammation by NRG4 appears to benefit insulin sensitivity in both prevention and treatment studies. It is known that chronic low-grade tissue inflammation is a major detrimental factor for obesity-associated metabolic disorders such as insulin resistance. Long-term nutrient overload in adipose tissue and the liver increases a stress response, leading to the recruitment and activation of macrophages and thus the activation of inflammatory pathways36. Consequently, insulin action is blunted in the stressed tissues by inhibiting insulin-signaling transduction, causing cellular insulin resistance37,38. Therefore, inhibiting inflammatory and macrophage infiltration by NRG4 contributes to improved insulin sensitivity in both the prevention and treatment studies. This conclusion is in agreement with the results of the studies where normal weight rats and db/db mice were used to assess whether acute injection of NRG1 protein improves glucose tolerance through the activation of ErbB3 receptors and phosphorylation of protein kinase B (PKB) and forkhead box protein O1(FOXO1) in the liver39,40.
In conclusion, the current study demonstrates that Nrg4 gene transfer inhibits lipogenesis and reduces chronic inflammation, resulting in the prevention of HFD-induced adiposity and fatty liver, and the improvement of insulin sensitivity. These findings provide direct evidence in support of the potential health benefits of NRG4 in managing obesity and obesity-associated metabolic disorders.
Materials and Methods
Materials
C57BL/6 mice were purchased from Charles River (Wilmington, MA). High-fat diet was obtained from Bio-serv (Frenchtown, NJ) (catalog number: F3282, 60% kJ/fat). The pLIVE and pLIVE-SEAP plasmids were purchased from Mirus Bio (Madison, WI). A Microvette-CB300-LH was from Fisher Scientific (Pittsburgh, PA). Thermocouple Meter was from Kent Scientific Corp (Torrington, CT). The insulin ELISA kit was obtained from Mercodia Developing Diagnostics (Winston Salem, NC). A TUREtrack glucometer and test strips were purchased from Nipro Diagnostics, Inc. (Fort Lauderdale, FL). The Oil Red O staining solution was obtained from Electron Microscopy Science (Hatfield, PA). The high-fidelity DNA polymerase was purchased from NEB (Ipswich, MA). The TRIZOL reagent and the SuperScript® III First-Strand Synthesis System were purchased from Life Technologies (Grand Island, NY). The RNeasy Lipid Tissue Mini Kit was from Qiagen (Valencia, CA). PerfeCTa® SYBR® Green FastMix was acquired from Quanta BioSciences (Gaithersburg, MD).
Plasmid construction
A coding region of mouse Nrg4 (NCBI GenBank, NM_032002) was cloned from complementary DNA sequences of C57BL/6 mouse liver and inserted into a pLIVE vector at the NheI and XhoI cutting sites. The insertion in the new plasmid (pLIVE-NRG4) was confirmed by DNA sequencing. The control plasmid pLIVE-SEAP contains the same backbone and a secreted alkaline phosphatase gene. Both pLIVE-NRG4 and pLIVE-SEAP plasmids were purified by CsCl–ethidium bromide density-gradient ultracentrifugation and kept in saline (0.9% sodium chloride). Purity of the plasmids was verified by absorbency ratio at 260 and 280 nm and by 1% agarose gel electrophoresis.
Animals and treatment
All procedures performed on animals were approved by the Institutional Animal Care and Use Committee at the University of Georgia, Athens, Georgia and the methods were carried out in accordance with the approved protocol #A2014-07-008-Y1-A0. For the prevention study, C57BL/6 mice (male, 8-week old) were housed under standard conditions with a 12 h light-dark cycle and fed an HFD for a total of 9 weeks. For the treatment study, mice were first fed an HFD for 25 weeks, reaching a body weight of 56 grams on average. Gene transfer was performed on the obese mice and the treated animals were continued on an HFD for additional 3 weeks. Hydrodynamic gene delivery was performed according to a pre-established procedure10,41,42. Briefly, an appropriate volume of saline solution (equivalent to 8% lean mass) containing 20 μg pLIVE-NRG4 or pLIVE-SEAP was injected through the tail vein within 5–8 sec. The body weight of each mouse was measured on an electronic balance, and body composition was analyzed using an EchoMRI-100TM (Echo Medical Systems, Houston, TX). Food intake per mouse was calculated based on the amount consumed divided by time and the number of mice per cage. Blood was collected at the desired time using a Microvette-CB300-LH. Rectal temperature of the mice was measured at the desired time using a specially designed Thermocouple Meter.
Histochemical analysis
After the mice were euthanized, the liver, EWAT, SubWAT, and BAT were collected and fixed in 10% formalin. Paraffined tissues were sectioned at a thickness of 6 μm and stained with H&E solution. For Oil Red O staining of liver sections, freshly collected liver samples were frozen in liquid nitrogen. Frozen sections (8 μm) were made and stained with 0.2% Oil Red O in 60% of isopropanol for 20 min and washed three times with phosphate buffered saline. A microscopic examination was performed and photographs were taken under a regular light microscope.
Glucose tolerance test (GTT) and insulin tolerance test (ITT)
The analyses were performed on week nine post gene transfer in the prevention study and on week three after gene transfer to obese mice in the treatment study. For GTT, mice were fasted for 6 h and injected intraperitoneally with glucose at 1.5 g/kg body weight. Blood samples were collected from the tail vein and glucose levels were measured using a glucometer. For ITT, mice were fasted for 4 h and intraperitoneally injected with insulin (0.75 U/kg, Eli Lilly Indianapolis, IN). Blood samples were collected from the tail vein and glucose levels were measured using a glucometer.
Gene expression analysis by real time PCR
Total RNA was isolated from the liver, pancreas, BAT, and EWAT using the TRIZOL reagent or an RNeasy kit. Two micrograms of total RNA were used for the first strand cDNA synthesis, as recommended by the manufacturer. Real time PCR was performed, using SYBR Green as an indicator, employing an ABI StepOne Plus Real Time PCR system. PCR was carried out for 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Fluorescence was read during the reaction, allowing a continuous monitoring of the amount of PCR product. The data were normalized using Gapdh mRNA as an internal control. The primer sequences employed are summarized in Supplemental Table S1.
Statistical analysis
A statistical analysis was performed using the Student’s t test. All data are reported as mean ± standard deviation (SD) with statistical significance set at P < 0.05.
Additional Information
How to cite this article: Ma, Y. et al. Preventing High Fat Diet-induced Obesity and Improving Insulin Sensitivity through Neuregulin 4 Gene Transfer. Sci. Rep. 6, 26242; doi: 10.1038/srep26242 (2016).
References
Harari, D. et al. Neuregulin-4: a novel growth factor that acts through the ErbB-4 receptor tyrosine kinase. Oncogene 18, 2681–2689 (1999).
Britsch, S. The neuregulin-I/ErbB signaling system in development and disease. Adv. Anat. Embryol. Cell Biol. 190, 1–65 (2007).
Bernard, J. K., McCann, S. P., Bhardwaj, V., Washington, M. K. & Frey, M. R. Neuregulin-4 is a survival factor for colon epithelial cells both in culture and in vivo . J. Biol. Chem. 287, 39850–39858 (2012).
McElroy, S. J. et al. The ErbB4 ligand neuregulin-4 protects against experimental necrotizing enterocolitis. Am. J. Pathol. 184, 2768–2778 (2014).
Hayes, N. V. et al. Expression of neuregulin 4 splice variants in normal human tissues and prostate cancer and their effects on cell motility. Endocr. Relat. Cancer 18, 39–49 (2011).
Dunn, M. et al. Co-expression of neuregulins 1, 2, 3 and 4 in human breast cancer. J. Pathol. 203, 672–680 (2004).
Nielsen, T. O., Friis-Hansen, L., Poulsen, S. S., Federspiel, B. & Sorensen, B. S. Expression of the EGF family in gastric cancer: downregulation of HER4 and its activating ligand NRG4. PLos One 9, e94606 doi: 10.1371/journal.pone.0094606 (2014).
Rosell, M. et al. Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am. J. Physiol. Endocrinol. Metab. 306, E945–964 (2014).
Wang, G. X. et al. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat. Med. 20, 1436–1443 (2014).
Liu, F., Song, Y. & Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6, 1258–1266 (1999).
Zhang, G., Budker, V. & Wolff, J. A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 10, 1735–1737 (1999).
Suda, T. & Liu, D. Hydrodynamic gene delivery: its principles and applications. Mol. Ther. 15, 2063–2069 (2007).
Zhang, G. et al. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 11, 675–682 (2004).
Suda, T. & Liu, D. Hydrodynamic delivery. Adv. Genet. 89, 89–111 (2015).
Oelkrug, R., Polymeropoulos, E. T. & Jastroch, M. Brown adipose tissue: physiological function and evolutionary significance. J. Comp. Physiol. B 185, 587–606 (2015).
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Fabbrini, E., Sullivan, S. & Klein, S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology 51, 679–689 (2010).
de Mutsert, R., Sun, Q., Willett, W. C., Hu, F. B. & van Dam, R. M. Overweight in early adulthood, adult weight change, and risk of type 2 diabetes, cardiovascular diseases, and certain cancers in men: a cohort study. Am. J. Epidemiol. 179, 1353–1365 (2014).
Anderson, J. W., Konz, E. C., Frederich, R. C. & Wood, C. L. Long-term weight-loss maintenance: a meta-analysis of US studies. Am. J. Clin. Nutr. 74, 579–584 (2001).
Kajimura, S., Spiegelman, B. M. & Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 22, 546–559 (2015).
Wang, G. X., Zhao, X. Y. & Lin, J. D. The brown fat secretome: metabolic functions beyond thermogenesis. Trends Endocrinol. Metab. 26, 231–237 (2015).
Townsend, K. L. & Tseng, Y. H. Brown fat fuel utilization and thermogenesis. Trends Endocrinol. Metab. 25, 168–177 (2014).
Balsamo, M. et al. Fauna Europaea: Gastrotricha. Biodivers Data J. e5800 (2015).
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin Invest. 109, 1125–1131 (2002).
Knebel, B. et al. Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass. PLos One 7, e31812 doi: 10.1371/journal.pone.0031812 (2012).
Shimomura, I., Bashmakov, Y. & Horton, J. D. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 274, 30028–30032 (1999).
Ma, Y. & Liu, D. Activation of pregnane X receptor by pregnenolone 16 alpha-carbonitrile prevents high-fat diet-induced obesity in AKR/J mice. PLos One 7, e38734 doi: 10.1371/journal.pone.0038734 (2012).
Strable, M. S. & Ntambi, J. M. Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit. Rev. Biochem. Mol. Biol. 45, 199–214 (2010).
Tang, J. J. et al. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 13, 44–56 (2011).
Fajas, L. et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J. Biol. Chem. 272, 18779–18789 (1997).
Koonen, D. P. et al. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes 56, 2863–2871 (2007).
Lee, Y. J. et al. Nuclear receptor PPAR gamma-regulated monoacylglycerol O-acyltransferase 1 (MGAT1) expression is responsible for the lipid accumulation in diet-induced hepatic steatosis. Proc. Natl. Acad. Sci. USA 109, 13656–13661 (2012).
Yu, S. et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J. Biol. Chem. 278, 498–505 (2003).
Ma, Y., Gao, M. & Liu, D. Chlorogenic acid improves high fat diet-induced hepatic steatosis and insulin resistance in mice. Pharm. Res. 32, 1200–1209 (2015).
Bu, L., Gao, M., Qu, S. & Liu, D. Intraperitoneal injection of clodronate liposomes eliminates visceral adipose macrophages and blocks high-fat diet-induced weight gain and development of insulin resistance. AAPS J. 15, 1001–1011 (2013).
Schenk, S., Saberi, M. & Olefsky, J. M. Insulin sensitivity: modulation by nutrients and inflammation. J. Clin. Inv. 118, 2992–3002 (2008).
Guo, J., Jou, W., Gavrilova, O. & Hall, K. D. Persistent diet-induced obesity in male C57BL/6 mice resulting from temporary obesigenic diets. Plos One 4, e5370 doi: 10.1371/journal.pone.0005370 (2009).
Lee, B. C. & Lee, J. Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance. Biochim. Biophys. Acta 1842, 446–462 (2014).
Ennequin, G. et al. Neuregulin 1 Improves Glucose Tolerance in db/db Mice. PLos One 10, e0130568 doi: 10.1371/journal.pone.0130568 (2015).
Caillaud, K. et al. Neuregulin 1 improves glucose tolerance in adult and old rats. Diabetes Metab. pii: S1262-3636(15)00114-7. doi: 10.1016/j.diabet.2015.08.003 (2015).
Ma, Y. & Liu, D. Hydrodynamic delivery of adiponectin and adiponectin receptor 2 gene blocks high-fat diet-induced obesity and insulin resistance. Gene Ther. 20, 846–852 (2013).
Ma, Y., Gao, M., Sun, H. & Liu, D. Interleukin-6 gene transfer reverses body weight gain and fatty liver in obese mice. Biochim. Biophys. Acta 1852, 1001–1011 (2015).
Acknowledgements
We would like to thank Mrs. Francisca Burnley for English editing. This work was supported in part by grants from NIH (RO1HL098295).
Author information
Authors and Affiliations
Contributions
Y.M. and D.L. designed experiments. Y.M. and M.G. conducted experiments. Y.M. and D.L. wrote the paper. All authors reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ma, Y., Gao, M. & Liu, D. Preventing High Fat Diet-induced Obesity and Improving Insulin Sensitivity through Neuregulin 4 Gene Transfer. Sci Rep 6, 26242 (2016). https://doi.org/10.1038/srep26242
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26242
This article is cited by
-
Correlation of asprosin and Nrg-4 with type 2 diabetes Mellitus Complicated with Coronary Heart Disease and the Diagnostic Value
BMC Endocrine Disorders (2023)
-
MiR-146a-5p, targeting ErbB4, promotes 3T3-L1 preadipocyte differentiation through the ERK1/2/PPAR-γ signaling pathway
Lipids in Health and Disease (2022)
-
Neuregulin 4 Attenuates Osteoarthritis Progression by Inhibiting Inflammation and Apoptosis of Chondrocytes in Mice
Calcified Tissue International (2022)
-
A Feedforward Loop within the Thyroid-Brown Fat Axis Facilitates Thermoregulation
Scientific Reports (2020)
-
Clinical significance of serum concentrations of neuregulin-4, in acute coronary syndrome
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.