Abstract
The genomes of Gram-negative bacteria encode paralogues and/or orthologues of global modulators. The nucleoid-associated H-NS and Hha proteins are an example: several enterobacteria such as Escherichia coli or Salmonella harbor H-NS, Hha and their corresponding paralogues, StpA and YdgT proteins, respectively. Remarkably, the genome of the pathogenic enteroaggregative E. coli strain 042 encodes, in addition to the hha and ydgT genes, two additional hha paralogues, hha2 and hha3. We show in this report that there exists a strong correlation between the presence of these paralogues and the virulence phenotype of several E. coli strains. hha2 and hha3 predominate in some groups of intestinal pathogenic E. coli strains (enteroaggregative and shiga toxin-producing isolates), as well as in the widely distributed extraintestinal ST131 isolates. Because of the relationship between the presence of hha2/hha3 and some virulence factors, we have been able to provide evidence for Hha2/Hha3 modulating the expression of the antigen 43 pathogenic determinants. We show that tracking global modulators or their paralogues/orthologues can be a new strategy to identify bacterial pathogenic clones and propose PCR amplification of hha2 and hha3 as a virulence indicator in environmental and clinical E. coli isolates.
Similar content being viewed by others
Introduction
Epidemiology of bacterial infections is in some instances understood because of the distribution of virulence genes in clinical isolates1. Eschericha coli virulent strains are a good example for that. This microorganism represents an outstanding example of genetic plasticity2 and of how the mechanisms driving horizontal gene transfer (HGT) impact its ability to colonize several niches, including human organs and tissues. Whereas several E. coli isolates are non-pathogenic and some of them belong to the human intestinal flora, many other strains express virulence determinants, which allow them to proliferate and cause disease. Pathogenic E. coli isolates are classified in pathotypes, which are defined by a combination of virulence factors, phenotype and clinical association3. However, the distribution of virulence factors is not strictly associated to each pathotype. A well-known example is E. coli strain O104:H4 that caused a large outbreak of bloody diarrhea with a high prevalence of associated hemolytic–uremic syndrome (HUS) in Germany in 20114. This newly emerged strain caused the highest frequency of HUS and death ever recorded. The O104:H4 outbreak strain was classified as an enteroaggregative E. coli (EAEC) because of its pattern of adherence to cultured cells and the presence of a plasmid (pAA) that encoded the fimbriae that mediate this type of adherence5. In contrast to typical EAEC strains, the outbreak strain contains a prophage encoding the Shiga toxin6, which is a well-studied virulence determinant usually expressed by a different E. coli pathotype, enterohemorrhagic E. coli (EHEC). Remarkably, the strain contains an unusual combination of genes that accounts for its pathogenicity and its extensive antibiotic resistance profile against a variety of beta-lactams5,7. Pathogenic bacterial isolates containing different combinations of genes justify that identification of specific pathogenic lineages may require a complex analysis of the presence of a large set of virulence traits.
In enteric bacteria, regulation of virulence determinants is dependent upon, among other global regulators, the nucleoid-associated protein H-NS. This protein is widespread in Gram-negative bacteria and has been best studied in E. coli and related genera. H-NS plays a dual role, both as an architectural protein that contributes to the nucleoid structure and as a global modulator of gene expression (for a review see8). The E. coli hns gene encodes a 137-amino-acid protein with a molecular mass of 15.4 kDa. H-NS binds to DNA in a non-sequence-specific manner, but with a preference for intrinsically curved AT-rich regions. Genome-wide ChIP- and microarray studies have identified the set of genes that are modulated by H-NS9. They are mainly located in HGT DNA and include several virulence determinants10,11,12.
The Hha family includes a group of sequence-related low molecular mass proteins (about 8 kDa) involved in gene regulation in the enterobacteria. These proteins show structural mimicry to the H-NS N-terminal domain and interact with this protein to modulate gene expression (as reviewed in13). The hha gene can be present in one or more copies per chromosome or in plasmids in members of the Enterobacteriaceae family, but not in other genera such as Vibrio or Aeromonas, which also express proteins of the H-NS family14. Studies on the mechanism of action of the Hha protein are based on the Hha-mediated down-regulation of the hlyCABD operon in E. coli, which encodes the toxin α-haemolysin. Instead of binding to specific regulatory sequences, Hha binds to H-NS, which in turn binds to specific regions of the hly operon15. Several Hha targets are HGT genes10,16, which include various virulence determinants from enteric bacteria, comodulated with H-NS14,17,18.
A general rule in several enterobacterial isolates as, for instance, Salmonella and E. coli strains, is the presence of both a paralogue of the hns and hha genes (the stpA and the ydgT genes respectively) in their genomes. In addition, orthologues of hns and hha are also encoded in several conjugative plasmids19. The role of H-NS and Hha paralogues is not yet well characterized. Both the StpA and YdgT proteins are overexpressed in mutants lacking either H-NS or Hha and, in these backgrounds, overexpression of the paralogues appears to attenuate the phenotype of either hns or hha mutants14,20. Other roles for both paralogues are not ruled out.
A recent genomic analysis performed by our group has shown that, unlike many other E. coli strains, the chromosome of the EAEC strain 04221 encodes four paralogues of the hha gene: hha, ydgT and the hitherto undescribed hha2 and hha3 alleles (Fig. 1). By studying their distribution among a large number of commensal and pathogenic E. coli strains, we provide in this report convincing evidence for the association of the presence of these allelles to highly virulent E. coli isolates. We also provide information about their biological role. We propose tracking alleles of global modulators as a new approach to identify and characterize pathogenic bacterial isolates.
Alignment of nucleotide (A) and aminoacid (B) sequences of Hha, Hha2 and Hha3. PCR amplification of the EC042_4516 (hha2) and EC042_4796 (hha3) genes (C) and hha, hha2, hha3 and ydgT genes (D) in strain 042 and different commensals E. coli strains. MW, molecular weight marker (GeneRuler 100 bp DNA Ladder, ThermoFisher Scientific); Lane 1, strain 042; lane 2, strain MG1655; lane 3, strain 5K; lane 4, strain XL1Blue; lane 5, strain BL21 (DE3); lane 6, strain DH5α; lane 7, strain ED1a; lane 8, strain ECO 01; lane 9, strain ECO 06; lane 10 negative PCR control.
Results
Identification of hha paralogues EC042_4516 and 4796 in the genome of the EAEC strain 042
hha paralogues EC042_4516 and EC042_4796 (from here on termed hha2 and hha3 respectively) were identified in the annotated genome of strain 042 by performing a Blast searching (http://www.uniprot.org/blast/) using the amino acid sequence of the Hha protein (Uniprot - D3GU89) as a template. Figure 1A,B show the nucleotide and aminoacid sequence alignment of Hha and putative Hha2 and Hha3 proteins. The ydgT gene was hitherto the unique chromosomally encoded hha paralogue described and can be detected in all E. coli isolates. We decided to use PCR to assess the distribution of Hha, YdgT and putative Hha2 and Hha3 proteins in a large number of E. coli isolates. Taking into account the high degree of similarity of hha, hha2 and hha3 genes, we designed primers that would specifically amplify hha2 and hha3 but not hha. To do this we performed a search for specific regions within hha2/hha3 paralogues by using the Primer Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Specific primers to amplify hha2 and hha3 genes (4516 forward/reverse and 4796 forward/reverse (Supplementary Table 1 and Supplementary Fig. 1)) were selected. To confirm their specificity, we used strains 042 (positive control) and the already sequenced MG1655, DH5α and BL21 E. coli strains (Table 1). In addition, the E. coli lab strain 5K and the commensal strain ED1a were also assayed. PCR analysis of the above-referred strains with the designed primers specific for hha2 and hha3 paralogues confirmed that they amplify these two hha alleles in strain 042, but not in the rest of E. coli strains analysed which, as expected, encode hha and ydgT (Fig. 1C,D).
Distribution of hha2 and hha3 genes among E. coli strains belonging to different pathotypes
The fact that hha2 and hha3 were not detected in the different E. coli laboratory strains tested led us to hypothesize that these genes might be predominantly encoded in pathogenic E. coli isolates. To assess their presence in the different E. coli pathotypes, we used a LREC collection of 60 strains that includes: 16 enteroaggregative E. coli (EAEC), 21 Shiga toxin (verotoxin)-producing E. coli (STEC/VTEC), 3 enterotoxigenic E. coli (ETEC), 2 typical enteropathogenic E. coli (tEPEC), 2 enteroinvasive E. coli (EIEC), 1 adherent-invasive E. coli (AIEC), 14 extraintestinal pathogenic E. coli (ExPEC) and 1 commensal isolate (Supplementary Table 2). Presence of hha2 and hha3 genes could be unambiguously assessed by PCR analysis using the above-indicated specific primers for these genes (Supplementary Fig. 2A,B). Interestingly, 62.5% of all EAEC isolates harbored either hha2 (62.5%) or hha3 (50%) alleles (Table 2). Distribution of hha2 and hha3 genes between STEC/VTEC strains was lower than those observed among EAEC isolates (43% for hha2 and 33% for hha3). With respect to ExPEC strains, the incidence of both paralogues is biased depending upon the sequence type: it is high among ST131 clone (67% for both genes) and low for the rest (38 and 13% for hha2 and hha3 respectively) (Table 2). It is important to point out here that all strains used in this study encode both hha and ydgT genes (Supplementary Fig. 3A,B).
Because of the high prevalence of hha2 and hha3 genes in the set of EAEC strains initially tested, we decided to perform a more comprehensive analysis of the presence of these paralogues in a larger number of EAEC isolates and to try to correlate it with the virulence factors expressed by these isolates (Supplementary Table 3). As expected from the preliminary analysis with 16 EAEC strains (Table 2), the analysis of a total number of 56 EAEC strains confirmed the prevalence of both alleles. Allele hha2 was present in 37 (66%) strains (in 10 of them alone, in the rest in combination with hha3). Allele hha3 was present in 29 strains (52%) (in 2 of them alone, in the rest in combination with hha2). Both alleles together are present in 27 strains (48%). Remarkably, all three O104:H4 EAEC strains analysed showed one (2 strains hha2) or both (1 strain) alleles and 5 of 6 O3:H2 EAEC strains were positive for one (hha2) or both alleles. In contrast, all five O86:H2 EAEC strains analysed did not show any of both paralogues.
The genes encoding the following virulence factors of EAEC were detected by PCR: Antiaggregation protein transporter (aatA gene), AAF/I fimbrial subunit (aggA gene), AAF/II fimbrial subunit (aafA gene), AAF/III fimbrial subunit (agg3A gene), transcriptional activator (aggR gene), aggregative heat-stable toxin 1 (EAST1) (astA gene), anti-aggregation protein (Dispersin) (aap gene), Shigella enterotoxin 1 mucinase (set1A gene), yersiniabactin (irp2 gene), serine protease Pet (pet gene), cryptic ORF (sfh gene), secreted autotransporter toxin (sat gene), serine protease Pic (pic gene) and antigen 43 (agn43 gene). The 14 virulence determinants studied are usually associated to EAEC strains, although sat is also associated to ExPEC strains and astA is associated to different pathotypes. From these genes, five (aggA, astA, shf, sat and agn43) were significantly associated with the presence of hha2 and hha3 alleles (see Supplementary Table 3).
To complete the analysis we examined a collection of 25 E. coli isolates from German humans and companion animals comprising mainly ExPEC22 (Supplementary Table 4). hha2 was present in 32% of the strains and hha3 in 56% of the strains (Table 3). Remarkably, these alleles predominate in ExPEC/EAEC strains and are very infrequent in those isolates considered as intestinal flora (Table 3). We took advantage of the fact that these strains have been sequenced to correlate PCR data with genomic data (Supplementary Table 5). There exists a good correlation between the PCR analysis and the in silico detection of hha2/hha3.
Distribution of hha2 and hha3 genes among the ECOR collection
We also decided to analyse the 72 members of the ECOR collection for the presence of hha2/hha3. The ECOR collection is a widely used set of 72 wild-type E. coli strains isolated between 1973 and 1983 from a variety of animal hosts and a variety of geographic locations23. The collection is thought to broadly represent genotypic variation in E. coli24. Although it was initially stated that none of the ECOR strains is pathogenic25, different reports have provided evidence for pathogenic E. coli being grouped among the ECOR strains on the basis of MLEE26. Furthermore, pathogenicity determinants for uropathogens, such as pap, hly, kps and sfa, are present in the genomes of some among of the ECOR strains27, though it is unclear whether or not these genes are active. Thus, 29 of 72 strains of ECOR collection showed the ExPEC status (with two or more of the following five virulence genes: pap, sfa/foc, afa, aer and kps)28 (Supplementary Table 6). PCR analysis of the distribution of hha2 and hha3 alleles showed that only 15% of the EcoR strains harbour any of them. Remarkably, both alleles were dectected in 4% of the strains only (Table 4).
Distribution of hha2 and hha3 genes in environmental E. coli isolates
To obtain a broader view about the distribution of both hha paralogues in E. coli, we decided to investigate their presence in the genomes of environmentally isolated E. coli strains. To perform this study we used firstly a collection of 84 environmental E. coli isolates that were selected because they encode the stx2 gene (Supplementary Table 7). These strains were isolated either from raw sewage samples of urban origin, mostly contaminated by human faecal wastes, or from wastewater samples from three different abattoirs (cattle, pig and a mixed cattle, lamb, goat slaughterhouse). The strains were isolated from the water samples only on the basis of the stx2 presence. Samples were isolated as previously described29. Secondly, we isolated a further set of 88 E. coli strains from environmental samples, which, without further characterization, were tested for the absence of stx and the presence of hha2 or hha3. In spite of their environmental origin, there was a significant bias in the distribution of the hha alleles in both sets of strains. hha2 alone was present in 35 out of the 84 Stx-producing E. coli strains (41.6%). hha3 alone was not present in any strain. Both alleles were present in six out of the 84 strains (7.1%) (Table 5). In contrast, when considering the 88 non-characterized Stx− environmental isolates, hha2 allele alone was present in ten strains (11.3%) (Significant differences versus Stx2+ strains (P = 0.000019)), hha3 alone was present in two strains (2.3%) and the combination of both in nine strains (10.2%).
Identification of hha2 and hha3 in the annotated Escherichia coli genomes
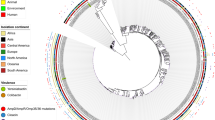
The annotated E. coli genomes in NCBI are grouped in 33 groups, each of which has a representative strain. We used the BLASTN (90% identity and 80% coverage) to detect hha2 and hha3 in 20 strains whose genome is complete. 16 strains correspond to representative strains of each group and 4 to members of groups whose representative strain has not been completely sequenced. hha2 and hha3 homologues could be identified in twelve strains (Fig. 2). Representative strain of group 1 is MG1655. As expected, any of both genes could be detected in this strain (data not shown). We extended our analysis to the rest of the strains of this group whose genome is completely sequenced. None of them expressed these paralogues. Identification of hha2 and hha3 was possible mainly in pathogenic E. coli representatives, out of the commensal strains SE15, O9H5 and ED1a, but this latter contains a 3 bp deletion of the hha2 homologue, which most likely results in function loss. We also mapped hha, ydgT, hha2 and hha3 genes in the corresponding genetic maps (Fig. 2). As expected from core genes, position of hha is similar in all genomes except that of strain O104:H4 (most likely this being due to a genomic rearrangement). This is also the case for ydgT. In contrast, hha2 and hha3 alleles map in different positions in the different strains that harbour them and they are flanked by insertion elements sequences. As well, hha2 and hha3 are distributed randomly in the different groups.
Map position of hha2 and hha3 genes in representatives of the different groups of NCBI annotated E. coli genomes.
In the top is drawn the genome of strain MG1655 including positions of hha and ydgT, which are fairly conserved in almost all E. coli genomes analysed. Thin line means hha2; thick line means hha3. Triangles correspond to transposase-like genes flanking either hha2 or hha3.
A regulatory role for the hha2 and hha3 gene products: modulation of the expression of the ag43 determinants of strain 042
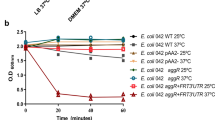
As commented above, the analysis of the presence of hha2 and hha3 genes in the set of EAEC strains analysed showed a correlation with the presence of, among others, the virulence factors shf and ag43. We decided to assess if any or both paralogues play a role modulating their expression. To evaluate this, we first constructed isogenic derivatives of strain 042 lacking either hha, hha2, hha3 or all three paralogues simultaneously. Analysis of the 042 genome annotation showed that this strain contains three different copies of the ag43 gene (EC042_2242, 4511 and 4803)30, but only one copy of the shf gene. We analysed the expression of the three ag43 copies and the shf gene in the different genetic backgrounds. Samples were obtained from cultures grown at 37 °C and entering the stationary phase (OD600 2.0). The analysis of the expression of the three different alleles of the ag43 gene was performed by qRT-PCR (Fig. 3A). For one of the ag43 alleles (EC042 _2242), all three hha paralogues modulate its expression. In contrast, expression of the other two is not significantly influenced by Hha or its paralogues (Fig. 3B). With respect to the shf gene, the results obtained show that the Hha protein represses its expression under the conditions tested (Fig. 3C). In this example, the other two hha paralogues do not appear to significantly participate in the modulation of shf.
(A) Relative expression of the three antigen 43 determinants encoded in E. coli strain 042. Expression levels of the different ag43 alleles in the hha and hha null mutants compared with those of the wt strain. (B) Expression of the antigen 43 (EC042_2242) in hha, hha2 and hha3 mutants compared with those of the wt strain. (C) Expression of the sfh gene in 042 hha and hha null strains, compared with that of the wt strain.
Comparative expression of hha, hha2 and hha3 alleles in strain 042
Expression of global modulators is a key aspect of their regulatory role. It is well known that proteins such as FIS show an increased expression in the exponential growth phase, other such as IHF are mainly expressed in the stationary growth phase31,32. Former studies showed that the Hha protein is reduced in salt-free LB medium33. We decided to compare expression of all three paralogues in strain 042. Samples were obtained from cultures grown at 37 °C to the early stationary phase (OD600 nm 2,0). Expression of all three paralogues was assessed by qRT-PCR (Fig. 4). hha2 expression is slightly lower than hha expression. Remarkably, expression of hha3 is significantly lower than those of the other two paralogues.
Discussion
Temporal and geographical surveillance of high-risk bacterial pathogenic clones is a relevant issue to prevent outbreaks. Identification of these clones is performed by a combination of both classical typing and subtyping protocols (i.e., detection of genes and/or gene products associated to virulence, serotyping, bacteriocin typing, antimicrobial testing and other phenotypic traits such as biochemical or enzymatic activities) with more recently developed genotyping approaches (i.e., pulse field electrophoresis, multilocus sequence typing or whole-genome sequencing (WGS)). WGS can be used for epidemiological applications (as reviewed in34), but also to define phenotypic characteristics (i.e., virulence) of a particular pathogen. Among these approaches, PCR has and is being used as a routinely tool to detect the distribution of virulence determinants that is frequently used to understand the epidemiology of bacterial infections1.
Due to the relevance of E. coli as an indicator of faecal contamination, there have been developed several methods for the detection of this microorganism in environmental as well as in food samples. In contrast to tedious and time-consuming classical methodologies such as the Multiple Tube Fermentation technique, approaches such as the PCR, are rapid, highly specific and sensitive35,36. Different genes, such as 16S rDNA, EF-Tu37, uidA or yaiO38 have been used to detect E. coli by PCR in environmental samples. For the detection of specific E. coli pathotypes, combinations of primers specific for virulence factors are designed39,40,41. Considering the high E. coli genetic plasticity and because of the impact of HGT shaping E. coli bacterial genome2, it is not surprising that isolates belonging to a specific E. coli pathotype may display unusual combinations of virulence factors. Even some of the hitherto considered commensal strains encode well-characterized virulence factors39,42,43,44. Hence, widely spread virulence markers different from classical virulence factors may help to identify pathogenic E. coli isolates.
Whereas several bacterial global modulators have been thoroughly studied in the recent past, this is not the case with their corresponding paralogues or orthologues. Plasmid-encoded orthologues of genes coding for nucleoid-associated proteins are not rare19 but again, their biological significance remains in many instances unclear. We show in this report that the study of paralogues or orthologues of global modulators may be of great relevance to better understand the biology of bacterial cells. In this context, the association of alleles of global modulators to specific E. coli pathotypes or highly virulent clones provides a new tool for the surveillance of pathogenic microorganisms. The presence of a global modulator must be necessarily associated to a set of genes rather than to a specific gene. These traits must in turn define a specific lifestyle of the isolate. Paralogues hha2 and hha3 match to this approach.
From the different sets of strains used for this study, hha2 and hha3 paralogues are mainly detected among EAEC, STEC/VTEC and ExPEC strains. With respect to this latter pathotype, it is remarkable that hha2 and hha3 are mainly associated to ST131 isolates. A correlation between hha2, hha3 and intestinal pathogenic strains is further established when environmental strains are selected because of the presence of the stx2 gene in their genomes. Whereas about 50% of the environmental strains encoding stx2 have incorporated hha2/hha3 alleles, their presence is only of about 20% when environmentally isolated E. coli strains lacking this virulence determinant are analysed. Analysis of the ECOR collection for the presence of the hha paralogues confirms the relationship between them and virulence. Within the ECOR strains, only of 15% encode hha2 and 4% hha3. The presence of hha2 and hha3, rather than being linked to a pathotype (i.e., EAEC), is a marker of a set of pathogenic isolates that includes clones causing some of the most severe E. coli-mediated infections (i.e., EAEC O104:H4 or the worldwide distributed ExPEC ST131 clone). PCR-mediated amplification of hha2/hha3 in environmental or clinical samples is therefore a preliminary indicator of the presence of highly virulent E. coli strains which, in turn, can be further characterized by using primers specific of virulence genes that are associated to specific pathotypes.
A variable mapping position, their genomic context and their random distribution in the E. coli phylogenetic groups strongly suggest hha2 and hha3 spreading in E. coli strains by horizontal gene transfer mechanisms. Interestingly, these alleles are not detected in plasmids, which speaks for hha2 hha3 spreading by mechanisms different to conjugation. It is reasonable to hypothesize that horizontal inheritance of hha2 and hha3 may be correlated with inheritance of other HGT-encoded genes, among others, virulence determinants.
In addition to the correlation between hha2, hha3 and virulent E. coli isolates we provide in this work insight about the biological role of their gene products. hha2 and hha3 expression is not significantly influenced by the growth conditions (i.e., exponential/stationary phase, high/low temperature, high/low osmolarity) (our unpublished results). By using as a model the EAEC strain 042, we show that, when growing in rich medium at 37 °C., modulation of the expression of one of the three copies of ag43 that are encoded in this strain requires the participation of the hha2 and hha3 gene products and of the Hha protein itself. This was not the case for the shf gene, which is present in a single copy and is modulated by the hha gene product, but not by its paralogues. Hence, a likely hypothesis that may at least in part explain the occurrence of these paralogues is that amplification of virulence determinants such as ag43 in some strains may positively select the amplification of the genes that encode for the global modulators that fine tune their expression. An intriguing aspect that deserves further research is the significant lower expression level of hha3 compared to hha and hha2.
Analysis of virulence in pathogenic microorganisms usually takes into account the presence or absence of the corresponding virulence gene(s). The results we provide here clearly show that it is relevant to consider also the presence of specific alleles of global modulators. The increasing availability of the complete genomic sequences of several pathogenic strains will facilitate this analysis and will provide a more complete view of the complexity of the virulence regulons that some bacterial pathogens display.
Methods
Bacterial strains, plasmids and culture media
All bacterial strains used in this work are listed in Table 1. Cultures were normally grown in Luria Broth (LB) medium (10 g NaCl, 10 g tryptone and 5 g yeast extract per litre) with vigorous shaking at 200 rpm (Innova 3100, New Brunswick Scientific).
Isolation of Stx− E. coli strains from water samples
E. coli isolation was performed using the membrane filtration method according to previously standardized methods45. Briefly, serial decimal dilutions of urban wastewater and river water were filtered through 0.44-mm-pore-diameter membrane filters (EZ-Pak® Membranes were placed on Chromocult® coliform agar (Merck, Darmstadt, Germany) for selective E. coli growth and incubated at 44 °C for 18 h. Blue colonies of each sample, corresponding to E. coli, were randomly subcultured and used for the study. Indol test was used to confirm that the isolates were E. coli and the absence of stx genes was confirmed by PCR.
Genetic manipulations
All enzymes used to perform standard molecular and genetic procedures were used according to the manufacturer’s recommendations. To introduce plasmids in E. coli, bacterial cells were grown until a D.O600 nm 0,6. Cells were then washed several times with glycerol 10% and the respective plasmids were introduced by electroporation using an Eppendorf gene pulser (Electroporator 2510).
Mutant derivatives lacking alleles hha, hha2 and hha3 in EAEC strain 042 were obtained by the λ Red recombinant method described by46. Briefly, the antibiotic-resistance cassette of kanamycin of plasmid pKD4 was amplified using oligonucleotides HhaP1/HhaP2, 4516P1/4526P2 and 4796P1/4796P2 for hha, hha2 and hha3 deletions, respectively (See Supplementary Table 1, for sequence). DNA templates were treated with DpnI (Thermo Scientific) following manufacturer recommendations, and, then, purified and electroporated to the competent cells. Mutants were selected on LB plates containing the appropriate selection marker (kanamycin in that case) and the successful deletion of the gene was confirmed by PCR using the primers KT (kanamycin resistance; Kmr) in combination with specific primers located in the remaining gene sequence in the bacterial chromosome.
If necessary, the antibiotic resistance was eliminated by transforming the mutant strain with plasmid pCP20 and subsequent incubation at 42 °C for two or more passages as reported47. The pCP20 plasmid encodes the Flp recombinase that catalyses the recombination between the FRT sites flanking the kanamycin cassette47.
Amplification of alleles hha2 and hha3 by PCR
To detect the prevalence of the different hha paralogues in the strains tested, we performed standard PCRs. One colony of each bacterial strain was diluted in 20 μl of sterile water and it was used as a template for the premix DreamTaq Green PCR Master Mix (Thermo Scientific), and, a final concentration of 0,4 μM of primers 4516FW–4516RV or 4796FW–4796RV, for paralogues hha2 and hha3, respectively (See supplementary Table 1, for sequence). PCRs were run using the following steps: initial denaturation for five minutes at 95 °C, following by 25 cycles of 95 °C denaturation temperature for 30 seconds, annealing temperature of 58 °C for 30 seconds and 30 seconds of 72 °C extension temperature followed by another ten minutes of final extension at 72 °C. The 25-cycle amplification was performed in a T100 thermal cycler (Biorad). For detection of PCR products, 10 μl of the amplified DNA was run on a 2% TAE 0,5×−agarose gel (Pronadisa), stained with ethidium bromide and visualized under UV light using Gel Doc XR+ system (Biorad).
Isolation of RNA
Total RNA was extracted from bacterial pellets using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instruction. Potential traces of DNA were removed by digestion with DNase I (Turbo DNA-free, Ambion), according to the manufacturer’s instructions. RNA concentration and RNA quality were measured using a Nano- Drop 1000 (Thermo Fisher Scientific).
qRT-PCR
Expression levels of hha paralogues and antigen 43 genes were analysed using real-time quantitative PCR. Briefly, 1 μg of total RNA isolated previously was reverse transcribed to generate cDNA using the High-capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s protocol. All samples within an experiment were reverse transcribed at the same time, the resulting cDNA diluted 1:100 in nuclease-free water and stored in aliquots at −80 °C until used. As a control, parallel samples were run in which reverse transcriptase was omitted from the reaction mixture.
Real-time PCR was carried out using SYBR green PCR master mix (Life Technologies) and the ABI Prism 7700 sequence detection system (Applied Biosystems). Specific oligonucleotides complementary to the genes of interest were designed using primer3 software. The primers were named 4516FW–4516RV or 4796FW–4796RV, for paralogues hha2 and hha3, respectively (see Supplementary Table 1). Relative quantification of gene expression of mutants versus wild type strain was performed using the comparative threshold cycle (CT) method. The relative amount of target cDNA was normalized using the gapA gene as an internal reference standard. Fold change values referring to relative expression of target genes in mutant strains versus wt strain were calculated by dividing the ΔCT (difference of Ct values between the target gene and the internal reference standard gapA gene) obtained for the different mutant strains versus wt strain.
Statistical analysis
Proportions were compared between groups by use of the Fisher’s exact test. P < 0.05 was considered to denote significant differences.
Additional Information
How to cite this article: Prieto, A. et al. Tracking bacterial virulence: global modulators as indicators. Sci. Rep. 6, 25973; doi: 10.1038/srep25973 (2016).
References
Sabharwal, N., Dhall, S., Chhibber, S. & Harjai, K. Molecular detection of virulence genes as markers in Pseudomonas aeruginosa isolated from urinary tract infections. Int J Mol Epidemiol Genet 5, 125–134 (2014).
Touchon, M. et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5, e1000344 (2009).
Croxen, M. A. et al. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880 (2013).
Frank, C. et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 365, 1771–1780 (2011).
Bielaszewska, M. et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11, 671–676 (2011).
Mayer, C. L., Leibowitz, C. S., Kurosawa, S. & Stearns-Kurosawa, D. J. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins (Basel) 4, 1261–1287 (2012).
Rohde, H. et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 365, 718–724 (2011).
Dorman, C. J. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2, 391–400 (2004).
Navarre, W. W. et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238 (2006).
Baños, R. C. et al. Differential Regulation of Horizontally Acquired and Core Genome Genes by the Bacterial Modulator H-NS. PLoS Genet 5, e1000513 (2009).
Tran, C. N. et al. A multifactor regulatory circuit involving H-NS, VirF and an antisense RNA modulates transcription of the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 39, 8122–8134 (2011).
Stratmann, T., Pul, U., Wurm, R., Wagner, R. & Schnetz, K. RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants. Mol. Microbiol. 83, 1109–1123 (2012).
Madrid, C., Balsalobre, C., García, J. & Juárez, A. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol. Microbiol. 63, 7–14 (2007).
Madrid, C., García, J., Pons, M. & Juárez, A. Molecular evolution of the H-NS protein: interaction with Hha-like proteins is restricted to enterobacteriaceae. J. Bacteriol. 189, 265–268 (2007).
Madrid, C. et al. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184, 5058–5066 (2002).
Vivero, A. et al. Modulation of horizontally acquired genes by the Hha-YdgT proteins in Salmonella enterica serovar Typhimurium. J. Bacteriol. 190, 1152–1156 (2008).
Ellison, D. W. & Miller, V. L. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 188, 5101–5112 (2006).
Queiroz, M. H., Madrid, C., Paytubi, S., Balsalobre, C. & Juárez, A. Integration host factor alleviates H-NS silencing of the Salmonella enterica serovar Typhimurium master regulator of SPI1, hilA. Microbiology (Reading, Engl.) 157, 2504–2514 (2011).
Shintani, M., Suzuki-Minakuchi, C. & Nojiri, H. Nucleoid-associated proteins encoded on plasmids: Occurrence and mode of function. Plasmid 80, 32–44 (2015).
Sonnenfield, J. M., Burns, C. M., Higgins, C. F. & Hinton, J. C. The nucleoid-associated protein StpA binds curved DNA, has a greater DNA-binding affinity than H-NS and is present in significant levels in hns mutants. Biochimie 83, 243–249 (2001).
Nataro, J. P. et al. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J. Infect. Dis. 171, 465–468 (1995).
Schmiedel, J. et al. Multiresistant extended-spectrum β-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol. 14, 187 (2014).
Ochman, H. & Selander, R. K. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157, 690–693 (1984).
Ochman, H. & Selander, R. K. Evidence for clonal population structure in Escherichia coli. Proc. Natl. Acad. Sci. USA 81, 198–201 (1984).
Milkman, R. & McKane, M. DNA sequence variation and recombination in E. coli. 127–142 (Symposia-Society for General Microbiology, 1995).
Pupo, G. M., Karaolis, D. K., Lan, R. & Reeves, P. R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65, 2685–2692 (1997).
Marklund, B. I. et al. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol. Microbiol. 6, 2225–2242 (1992).
Clermont, O. et al. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11, 654–662 (2011).
Blanch, A. R., García-Aljaro, C., Muniesa, M. & Jofre, J. Detection, enumeration and isolation of strains carrying the stx2 gene from urban sewage. Water Sci. Technol. 47, 109–116 (2003).
Chaudhuri, R. R. et al. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS ONE 5, e8801 (2010).
Ball, C. A., Osuna, R., Ferguson, K. C. & Johnson, R. C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 174, 8043–8056 (1992).
Ditto, M. D., Roberts, D. & Weisberg, R. A. Growth phase variation of integration host factor level in Escherichia coli. J. Bacteriol. 176, 3738–3748 (1994).
Mouriño, M. et al. Osmolarity modulates the expression of the Hha protein from Escherichia coli. FEMS Microbiol. Lett. 160, 225–229 (1998).
Sabat, A. J., Budimir, A., Nashev, D. & Sá-Leão, R. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 18, Issue 4, 20380, (2013).
Buckalew, D. W., Hartman, L. J., Grimsley, G. A., Martin, A. E. & Register, K. M. A long-term study comparing membrane filtration with Colilert defined substrates in detecting fecal coliforms and Escherichia coli in natural waters. J. Environ. Manage. 80, 191–197 (2006).
Paton, A. W. & Paton, J. C. Detection and characterization of STEC in stool samples using PCR. Methods Mol. Med. 73, 45–54 (2003).
Molaee, N., Abtahi, H., Ghannadzadeh, M. J., Karimi, M. & Ghaznavi-Rad, E. Application of Reverse Transcriptase -PCR (RT-PCR) for rapid detection of viable Escherichia coli in drinking water samples. J Environ Health Sci Eng 13, 24 (2015).
Molina, F. et al. Improved detection of Escherichia coli and coliform bacteria by multiplex PCR. BMC Biotechnol. 15, 48 (2015).
Wu, X.-Y. et al. Comparative analysis of virulence genes, genetic diversity and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl. Environ. Microbiol. 73, 83–91 (2007).
Ndlovu, T., Le Roux, M., Khan, W. & Khan, S. Co-detection of virulent Escherichia coli genes in surface water sources. PLoS ONE 10, e0116808 (2015).
Omar, K. B. & Barnard, T. G. Detection of diarrhoeagenic Escherichia coli in clinical and environmental water sources in South Africa using single-step 11-gene m-PCR. World J. Microbiol. Biotechnol. 30, 2663–2671 (2014).
Calhau, V., Ribeiro, G., Mendonça, N. & Da Silva, G. J. Prevalent combination of virulence and plasmidic-encoded resistance in ST 131 Escherichia coli strains. Virulence 4, 726–729 (2013).
Sahl, J. W. et al. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect. Immun. 79, 950–960 (2011).
Qin, X. et al. Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains. PLoS ONE 8, e61169 (2013).
Way, C. Standard methods for the examination of water and wastewater. Water Environment Federation (2012).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Cherepanov, P. P. & W, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 (1995).
Guyer, M. S., Reed, R. R., Steitz, J. A. & Low, K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb. Symp. Quant. Biol. 45 Pt 1, 135–140 (1981).
Nieto, J. M., Prenafeta, A., Miquelay, E., Torrades, S. & Juárez, A. Sequence, identification and effect on conjugation of the rmoA gene of plasmid R100-1. FEMS Microbiol. Lett. 169, 59–66 (1998).
Taylor, R. G., Walker, D. C. & McInnes, R. R. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21, 1677–1678 (1993).
Bullock, W. O., Fernandez, J. M. & Short, J. M. XL1-Blue: A high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques 5, 376–379 (1987).
Studier, F. W. & Moffatt, B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 (1986).
Martínez-Castillo, A. et al. Type III effector genes and other virulence factors of Shiga toxin-encoding Escherichia coli isolated from wastewater. Environ Microbiol Rep 4, 147–155 (2012).
Acknowledgements
The authors acknowledge funding from the Spanish MICINN-FEDER (BFU2010-21836-C02-01) and Mineco (BIO2013-49148-C2-1-R and BIO2015-69085-REDC). Work in the LREC-USC-laboratory was financed by the grant CN2012/303 from Consellería de Cultura, Educación e Ordenación Universitaria (Xunta de Galicia) and The European Regional Development Fund (ERDF). M. Bosch is acknowledged for her contribution to the in silico analysis.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: A.P., M.H. and A.J. Performed the experiments: A.P., I.U., G.D., P.Q. and I.F. Analysed the data: A.P., J.B., M.T.M., T.C., M.H. and A.J. Wrote the manuscript: J.B., M.T.M., L.F., T.C., M.H. and A.J. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Prieto, A., Urcola, I., Blanco, J. et al. Tracking bacterial virulence: global modulators as indicators. Sci Rep 6, 25973 (2016). https://doi.org/10.1038/srep25973
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep25973
This article is cited by
-
Gene duplications in the E. coli genome: common themes among pathotypes
BMC Genomics (2019)
-
Extensive inter-strain diversity among clinical isolates of Shigella flexneri with reference to its serotype, virulence traits and plasmid incompatibility types, a study from south India over a 6-year period
Gut Pathogens (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.