Abstract
Salmonella typhimurium is a major cause of diarrhea and causes significant morbidity and mortality worldwide and perturbations of the gut microbiota are known to increase susceptibility to enteric infections. The purpose of this study was to investigate whether a Microbial Ecosystem Therapeutic (MET-1) consisting of 33 bacterial strains, isolated from human stool and previously used to cure patients with recurrent Clostridium difficile infection, could also protect against S. typhimurium disease. C57BL/6 mice were pretreated with streptomycin prior to receiving MET-1 or control, then gavaged with S. typhimurium. Weight loss, serum cytokine levels and S. typhimurium splenic translocation were measured. NF-κB nuclear staining, neutrophil accumulation and localization of tight junction proteins (claudin-1, ZO-1) were visualized by immunofluorescence. Infected mice receiving MET-1 lost less weight, had reduced serum cytokines, reduced NF-κB nuclear staining and decreased neutrophil infiltration in the cecum. MET-1 also preserved cecum tight junction protein expression and reduced S. typhimurium translocation to the spleen. Notably, MET-1 did not decrease CFUs of Salmonella in the intestine. MET-1 may attenuate systemic infection by preserving tight junctions, thereby inhibiting S. typhimurium from gaining access to the systemic circulation. We conclude that MET-1 may be protective against enteric infections besides C. difficile infection.
Similar content being viewed by others
Introduction
The commensal bacteria of the gastrointestinal (GI) tract play an important role in maintaining normal barrier function and intestinal homeostasis1,2,3 and in protecting against pathogenic infections4. In addition to protecting the host from colonization by newly ingested microorganisms (colonization resistance), these microbes have direct effects on host inflammatory pathways and barrier function2,3,5. Shifts in the composition of the gut microbiota resulting in an imbalance (‘dysbiosis’) may affect normal function and contribute to diseases such as irritable bowel syndrome6,7, inflammatory bowel disease8,9, obesity10,11 and diabetes12,13,14.
Dysbiosis can also increase susceptibility to enteric infections15,16. Salmonella enterica serovar Typhimurium (S. typhimurium) is an important human pathogen and major cause of diarrhea, causing over 93.8 million cases of gastroenteritis and 155,000 deaths per year17. Prior antibiotic use has been linked to Salmonella infection in humans and pre-exposure to antibiotics, particularly within two weeks prior to Salmonella infection, increases the risk of invasive non-typhoidal Salmonella disease in previously healthy children18,19. Animal studies of S. typhimurium infection suggest that the composition of the gut microbiota is important in protection4,20. Mice given the antibiotic streptomycin prior to S. typhimurium ingestion develop acute inflammation of the gastrointestinal (GI) tract with weight loss, diarrhea and tissue inflammation localized to the cecum, as well as systemic inflammation and extra-intestinal spread of infection to other tissues (e.g. spleen) at later time points21,22. Moreover, S. typhimurium-infected mice colonized with a less diverse defined minimal microbiota composed of 8 bacterial species (Altered Schaedler Flora) were colonized by S. typhimurium at high levels (108 CFU/g) and elicited pronounced enteropathogenesis by two days post-infection4.
Despite growing recognition that the microbiota plays an important role in protection against enteric infections, the underlying mechanisms remain unclear. We have developed a Microbial Ecosystem Therapeutic (MET-1) which has been used previously to cure recurrent Clostridium difficile disease in humans23. MET-1 is a defined, gut-derived microbial ecosystem of 33 bacteria cultured from the feces of a healthy human volunteer. As with S. typhimurium, Clostridium difficile disease is also associated with prior antibiotic exposure. The present study sought to expand our understanding of the function of MET-1 by investigating whether pretreatment with MET-1 could protect against disease in an antibiotic-associated murine model of S. typhimurium infection. We hypothesized that MET-1 would attenuate S. typhimurium infection through effects on the host barrier function, as well as on the pathogen itself.
Results
To demonstrate the protective effect of MET-1, we used the S. typhimurium colitis model in which C57BL/6 mice were gavaged with 20 mg of streptomycin following a 4 hour fast then after an additional 18 hours were gavaged with MET-1, or vehicle control (VC). Four hours later, mice were gavaged with either S. typhimurium (“infected”) or phosphate-buffered saline (“uninfected”). Mice were euthanized 48 hours later and samples (tissues and blood) collected for analysis. (Note: in the figures, VC indicates mice that have been treated with vehicle control and streptomycin).
MET-1 attenuates weight loss in Salmonella typhimurium-infected mice
To determine whether MET-1 administration could confer protection against S. typhimurium infection, we first measured mouse body weight daily as a general indicator of the overall health of each animal21. S. typhimurium-infected mice (VC + S. Tm. mice) lost significant weight compared to uninfected controls (Fig. 1a). However, infected mice receiving MET-1 lost significantly less weight than mice gavaged with vehicle control (−3.50% vs. −6.83%, respectively; p < 0.05). Mice not infected with S. typhimurium demonstrated normal weight gain.
MET-1 attenuated systemic markers of disease in S. typhimurium-infected mice.
(a) MET-1 attenuated weight loss in S. typhimurium infected mice. Following oral infection with S. typhimurium, mice were weighed daily and the percent change in weight from 0 to 48 hours is shown. VC = uninfected mice pretreated with vehicle control, n = 16; MET-1 = uninfected mice pretreated with MET-1, n = 16; VC + S. Tm. = S. typhimurium-infected mice pretreated with vehicle control, n = 18; MET-1 + S. Tm. = S. typhimurium-infected mice pretreated with MET-1, n = 18. Data were analyzed with a 2-way ANOVA using Bonferroni correction (*p < 0.05 at 48 hours), NS = not significant.
MET-1 inhibits Salmonella typhimurium growth in vitro but not in the intestine
Studies showing that the microbiota can inhibit growth of pathogens4,24 suggest that microbe-to-microbe interactions might decrease the burden of Salmonella in infected animals receiving MET-1. In vitro time kill assays of S. typhimurium with MET-1 indicated that MET-1 was able to inhibit growth of S. typhimurium, although MET-1 was unable to completely kill S. typhimurium at the concentrations tested (Fig. 2a). In contrast, in vivo we found no evidence that MET-1 + S. Tm. mice had a lower colonic pathogen burden compared to VC + S. Tm. mice (Fig. 2b). Similar results were noted in the stool (p > 0.05, data not shown). Analysis of mouse stool by 16S rRNA gene sequencing of samples taken 48 hours post-infection comparing the data from 5 separate experiments run several weeks apart showed clustering of samples was not based on the different groups the starting microbiota in each group had little effect on the course of treatment (Figure S1). Therefore, it is unlikely that direct killing of S. typhimurium by MET-1 is involved in either decreased systemic inflammation or decreased pathogen translocation.
MET-1 inhibited S. typhimurium in vitro but did not inhibit its growth in vivo.
(a) MET-1 (1:2, 1:100 and 1:300 dilutions) was incubated with S. typhimurium (S. Tm.), aliquots were withdrawn at times indicated, plated, counted and plotted as log10 CFU. MET-1 inhibited the growth of S. Tm. relative to the S. Tm. control, although MET-1 was unable to completely kill S. Tm. even at a high (1:2) concentration. (b) Colons were harvested 48 hours after S. Tm. infection, homogenized in sterile PBS and plated. Infected mice pretreated with MET-1 (MET-1 + S. Tm.) showed no significant reduction of S. Tm. counts in the colon (p > 0.05) compared to mice pretreated with vehicle control (VC + S. Tm.). S. Tm. was not detected in the colon of uninfected mice. Data were analyzed using a 1-way ANOVA with Bonferroni correction. VC = uninfected mice pretreated with vehicle control, n = 18; MET-1 = uninfected mice pretreated with MET-1, n = 18; VC + S. Tm. = S. typhimurium-infected mice pretreated with vehicle control, n = 22; MET-1 + S. Tm. = S. typhimurium-infected mice pretreated with MET-1, n = 21.
MET-1 did not decrease intracellular invasion of intestinal epithelial cells by Salmonella typhimurium
A commonly described mechanism whereby S. typhimurium causes infection involves active invasion of host cells25. This mechanism is independent of tight junction protein function and involves invasion of host cells via the Type III Secretion System (TTSS) of S. typhimurium to inject bacterial effector proteins, ultimately leading to internalization of the bacteria into the host cell25. Although major growth inhibition of Salmonella by MET-1 was not observed in Fig. 2, it has been reported that Clostridial species present in the fecal microbiota can inhibit the invasion virulence properties of Salmonella typhimurium26. To determine whether MET-1 could protect against inhibition of host cell invasion by S. typhimurium, or otherwise affect the invasive virulence capacity of S. typhimurium, we used an in vitro cell culture gentamicin-based invasion assay to determine the effect of MET-1 pretreatment on the susceptibility of Caco-2 intestinal epithelial cells to bacterial invasion by S. typhimurium. No differences were noted in the CFU/mL of intracellular S. typhimurium in either the MET-1 or saline groups treated with S. typhimurium (p > 0.05), indicating that bacterial invasion of intestinal epithelial cells was not prevented by MET-1 pretreatment and MET-1 did not result in statistically lower counts of Salmonella bacteria in this assay (Fig. 3). The controls of uninfected cell monolayer pretreated with saline, MET-1, or heat-killed S. typhimurium plus MET-1 did not result in any bacterial growth. These data suggest that MET-1 does not significantly affect the virulence or invasive capacity of S. typhimurium.
MET-1 pre-treatment does not prevent S. typhimurium intracellular invasion of Caco-2 epithelial cells.
Live (or heat-killed) S. typhimurium were added to Caco-2 cell monolayers at an MOI of 100:1 and incubated for 1 hour. Extracellular bacteria were then killed by the addition of gentamicin (100 μg/mL). One hour later, Caco-2 cells were lysed and intracellular bacteria were enumerated using serial dilutions plated on MacConkey agar plates containing 100 μg/mL streptomycin. There were no significant differences between S. typhimurium-infected cells pretreated with MET-1 (MET-1 + Sal) or saline (Saline + Sal) (p > 0.05). No bacterial growth was seen in cell monolayers treated with saline only (Saline), MET-1 only (MET-1), heat-killed S. typhimurium only (Saline + HK Sal) or MET-1 pretreated cell monolayers treated with heat-killed S. typhimurium (MET-1 + HK Sal). Data were analyzed using a 1-way ANOVA with Bonferroni correction, n = 3, (p > 0.05) NS = not significant.
MET-1 reduced systemic spread of infection
Since the ability of S. typhimurium to translocate from the gut and cause metastatic infection is well described21, we sought to determine whether MET-1 would affect the ability of S. typhimurium to disseminate to other organs. Quantification of S. typhimurium in the spleen was determined by measuring the number of CFU/g of tissue and bacterial speciation was confirmed by PCR. MET-1 + S. Tm mice had significantly lower S. typhimurium counts within the spleen compared with VC + S. Tm. mice (Fig. 4), indicating that MET-1 administration attenuated the ability of S. typhimurium to gain access to the systemic circulation. None of the uninfected mice had S. typhimurium present in the spleen.
Translocation of S. typhimurium to the spleen was reduced in MET-1 pretreated mice.
Spleens were harvested 48 hours after S. typhimurium infection, weighed and homogenized in 1 mL PBS. Serial dilutions were plated on MacConkey agar plates containing 100 μg/mL streptomycin. Plates were incubated at 37 °C for 24 hrs and the resulting colonies were counted. Infected mice pretreated with MET-1 (MET-1 + S. Tm., n = 13) had reduced S. typhimurium counts in the spleen (*p < 0.05) compared to mice pretreated with vehicle control (VC + S. Tm., n = 12). S. typhimurium was not detected in the tissues of uninfected mice pretreated with vehicle control (VC, n = 12) or MET-1 (n = 12). Data were analyzed using a 1-way ANOVA with Bonferroni correction.
MET-1 attenuates the increase in serum cytokine levels caused by Salmonella typhimurium infection
We next investigated whether MET-1 might attenuate the increase in systemic cytokine levels that occur after oral S. typhimurium infection27. By forty-eight hours, significant differences were seen for TNF-α, IL-1β, IL-12 p40, IL-12 p70, GM-CSF, eotaxin, MIP-1β and MCP-1 (Fig. 5a). All uninfected mice (VC mice and MET-1 mice) had similar levels of serum cytokines (p > 0.05, data not shown), while addition of MET-1 significantly reduced circulating levels of chemoattractants and inflammatory cytokines that are normally elevated in S. typhimurium infection. Chemoattractants for immune cells, particularly neutrophils28, were found at higher levels in the systemic circulation of the VC + S. Tm. mice compared to MET-1 + S. Tm. mice (Fig. 5a), suggesting that a key aspect of MET-1 action might include modulation of neutrophil mobilization.
Pro-inflammatory cytokine levels were reduced in S. typhimurium-infected mice pretreated with MET-1.
(a) Serum cytokine levels were measured 48 hours post-infection using a Bio-Plex Pro mouse cytokine magnetic bead kit. Of the cytokines measured, TNF-α, IL-1β, IL-12p40, IL-12p70, GM-CSF, eotaxin, MIP-1β and MCP-1 were all significantly reduced in infected mice pretreated with MET-1 (MET-1 + S. Tm.) compared to infected mice pretreated with vehicle control (VC + S. Tm.). There were no significant differences in cytokine concentrations between uninfected mice pretreated with MET-1 or vehicle control (data not shown). Data were analyzed using a 1-way ANOVA with Bonferroni correction, n = 7 for each group (*p < 0.05, **p < 0.01, ***p < 0.001). (b) Cecal TNF-α expression was attenuated in S. typhimurium-infected mice pretreated with MET-1, as determined by quantitative RT-PCR (qRT-PCR) was performed in triplicate. The relative TNF-α expression was normalized to an endogenous control (GAPDH expression). The relative expression of TNF-α was attenuated in infected mice pretreated with MET-1 (MET-1 + S. Tm.) compared to infected mice pretreated with vehicle control (VC + S. Tm.). The level of expression in MET-1 + S. Tm. mice was not significantly different from uninfected mice pretreated with vehicle control (VC) or MET-1. Data were analyzed using a 1-way ANOVA with Newman-Keuls multiple comparison test, n = 4 for each group (*p < 0.05).
MET-1 attenuates local TNF-α expression induced by Salmonella typhimurium infection in cecum
To determine whether MET-1 also affected local pro-inflammatory cytokine expression in the intestine, we next looked at the expression of TNF-α in the cecum using quantitative real-time PCR. TNF-α is a key mediator of inflammation in the early stages of Salmonella infection and acts by recruiting neutrophils to the site of infection29,30. It was found that the expression of TNF-α was significantly increased in VC + S. Tm. mice compared to uninfected controls (Fig. 5b). However, MET-1 effectively reduced TNF-α expression in the MET-1 + S. Tm. treatment group. The addition of MET-1 alone had no significant effect on TNF-α expression.
MET-1 attenuates local neutrophil infiltration in cecum induced by Salmonella typhimurium infection
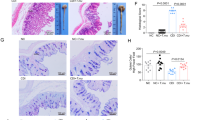
Localized responses of the immune system were examined in the intestinal mucosa, where infection with S. typhimurium causes an influx of neutrophils31. At 48 hours post-infection, immunocytochemistry showed increased staining for cecum MPO, a principal marker of neutrophils32, which was significantly reduced by treatment with MET-1 (Fig. 6a). Image analysis and quantification showed a 5-fold reduction in MPO staining in MET-1 + S. Tm. mice versus VC + S. Tm. mice (Fig. 6b). In addition, the nuclear localization of NF-κB, a key transcription factor and regulator of pro-inflammatory pathways that translocates to the nucleus when activated, was found to be reduced (Fig. 6c). The addition of MET-1 alone had no significant effect on either nuclear NF-κB or neutrophil accumulation in the ceca of uninfected mice.
MET-1 attenuates local neutrophil infiltration induced by S. typhimurium infection in cecum (a) Representative images of myeloperoxidase (MPO) immunofluorescence staining for neutrophils (green) in ceca. The area in the box (left) is displayed at a higher magnification in the panels on the right. Nucleic acid was stained using Hoechst (blue). S. typhimurium-infected mice pretreated with vehicle control (VC + S. Tm.) had more abundant immunostaining in the ceca compared to uninfected controls (VC) and was significantly reduced in the ceca of infected mice pretreated with MET-1 (MET-1 + S. Tm. mice). No MPO staining was noted in secondary antibody control slides for each group (not shown). (b) Quantification of MPO immunostaining, shown by the ratio of MPO (green immunofluorescence) staining to number of cells (blue nuclei). Data analyzed using 1-way ANOVA with Bonferroni correction, ***p < 0.001. n = 4 for VC, MET-1 and MET-1 + S. Tm. groups, n = 3 for VC + S. Tm. group. 3–5 fields of view per section. (c) Representative images of NF-κB P65 immunofluorescence staining (red) in ceca. The area in the box (left) is displayed at a higher magnification in the panels on the right. Nuclei were stained using Hoechst (blue). VC + S. Tm. mice had significantly increased NFκB P65 translocation (pink immunofluorescence) into the nucleus compared with MET-1 + S. Tm. mice. Secondary antibody control slides for each group showed no NFκB P65 staining (images not shown). (d) Ratio of NF-κB P65 nuclear staining to the total number of epithelial cells per high power field. The percentage of NF-κB P65 stained cells was normalized to the VC + S. Tm. group. Images taken at 400× magnification from at least two sections per cecum tissue. At least five non-adjacent images were taken for each section. Images were quantified from two independent experiments and each experiment had at least three mice per group. Total number of mice for each treatment group for two independent experiments was 6- to 9. Data were analyzed using ANOVA with Bonferroni correction, (***p < 0.001 and *p < 0.05).
MET-1 does not enhance intestinal mucin production in cecum
Since MUC2 has been proposed as an important host defense mechanism against S. typhimurium33,34, mucin production was examined in VC + S. Tm. mice and MET-1 + S. Tm. mice. Alcian blue staining showed that MET-1 + S. Tm. mice did not have increased staining (Fig. 7a) or goblet cell counts (Fig. 7b) compared to VC + S. Tm. mice. Moreover, no differences in MUC2 immunostaining were noted in MET-1 + S. Tm. mice compared to VC + S. Tm. mice (Fig. 7c,d). These data argue against MET-1-induced differences in intestinal mucus as a likely mechanism for conferring systemic protection against S. typhimurium.
MET-1 did not enhance intestinal mucin production in ceca of S. typhimurium- infected mice.
(a) Alcian blue staining (blue) of ceca fixed in 10% formalin were counterstained with nuclear red fast solution (red). (b) Goblet cells were enumerated from 10 random high-powered field views (400× magnification) spanning from muscularis mucosa to surface epithelium on Alcian blue stained sections. Infected mice pretreated with vehicle control (VC + S. Tm.) had less mucin staining than uninfected mice pretreated with vehicle control (VC + S. Tm.). However, there was no statistically significant difference between infected mice pretreated with MET-1 (MET-1 + S. Tm.) and vehicle control (VC + S. Tm.). (c) MUC2 immunofluorescence staining (green) in ceca fixed in Methanol-Carnoy’s solution. Nucleic acid was stained using DAPI (blue). Uninfected mice pretreated with VC or MET-1 had similar localization of MUC2 staining that differed from VC + S. Tm. mice. Again, MUC2 staining in MET-1 + S. Tm. mice more closely resembled VC + S. Tm. mice than uninfected controls. Quantification shown in (d). Data were analyzed using a 1-way ANOVA with Bonferroni correction. n = 3, (**p < 0.01, ***p < 0.001) NS = not significant.
MET-1 attenuates disruption of tight junction proteins in cecum caused by Salmonella typhimurium
S. typhimurium causes compromise of the intestinal epithelial barrier via disruption of tight junction proteins including ZO-1 and claudin 125,35. Bacterial entry through the paracellular route, in addition to direct cellular invasion35, is thought to be an important contributor to dissemination of infection36. In uninfected mouse ceca (VC mice or MET-1 mice), ZO-1 immunofluorescence was abundant in epithelial cells (Fig. 8a). In contrast, VC + S. Tm. mice had loss of ZO-1 membrane staining which was preserved in MET-1 + S. Tm. mice. Western blot analysis of the membrane-associated fraction of intestinal lysates demonstrated increased expression of ZO-1 in MET-1 + S. Tm. mice compared to VC + S. Tm. mice (Fig. 8b), indicating preserved expression of ZO-1 in MET-1 + S. Tm. mice.
MET-1 pretreatment prevented the disruption of ZO-1 and claudin-1 cellular localization in the cecum caused by S. typhimurium.
(a) ZO-1 immunofluorescence staining (green) in ceca fixed in 10% formalin (400× magnification). Nuclei were stained using Hoechst (blue). White arrows indicate green staining of ZO-1 protein at the membrane of intestinal epithelial cells. VC = uninfected mice pretreated with vehicle control; MET-1 = uninfected mice pretreated with MET-1; VC + S. Tm. = S. typhimurium-infected mice pretreated with vehicle control; MET-1 + S. Tm. = S. typhimurium-infected mice pretreated with MET-1. VC + S. Tm. mice had a loss of ZO-1 membrane staining that was attenuated in MET-1 + S. Tm. mice. (b) Expression of ZO-1 measured by Western blot analysis (S, Triton X-soluble fraction; I, Triton X-insoluble fraction, sample blot shown in top right of panel). Consistent with data in panel (a), densitometric values calculated for the ratio of ZO-1 to β-actin showed that MET-1 + S. Tm. mice had increased expression of ZO-1 in insoluble fractions compared to VC + S. Tm. mice. Data were analyzed using a 1-way ANOVA with Bonferroni correction (*p < 0.05). n = 5 mice for VC + S. Tm., n = 4 for MET-1 + S. Tm. (c) Claudin-1 immunofluorescence staining (green) in ceca fixed in 10% formalin (400x magnification). Nucleic acid was stained using Hoechst (blue). VC + S. Tm. mice displayed disaggregation and clumping of epithelial claudin-1 immunofluorescence, along with discontinuities in membrane staining and a reduction in staining intensity. This alteration in claudin-1 expression was attenuated in MET-1 + S. Tm. mice. (d) Expression of claudin-1 was measured by Western blot analysis (S, Triton X-soluble fraction; I, Triton X-insoluble fraction, sample blot shown). Densitometric values were calculated for the ratio of claudin-1 to actin and showed that MET-1 + S. Tm. mice had increased expression of claudin-1 in insoluble fractions compared to VC + S. Tm. mice. Data were analyzed using a 1-way ANOVA with Bonferroni correction (**p < 0.01). n = 5 mice for VC + S. Tm., n = 4 for MET-1 + S. Tm.
Similarly, VC + S. Tm. mice showed reduced staining intensity, as well as disaggregation and clumping of epithelial claudin-1 immunofluorescence. This outcome was markedly attenuated in MET-1 + S. Tm. Mice (Fig. 8c,d). Additionally, Western blot analysis showed that MET-1 + S. Tm. mice had increased claudin-1 expression in the membrane fractions compared to VC + S. Tm. mice (Fig. 8d), showing that administration of MET-1 in S. typhimurium-infected mice preserved claudin-1 expression.
Discussion
This study sought to address whether a defined gut microbial community that has been previously used to cure C. difficile infection23 may also act to protect against S. typhimurium, another gut pathogen and important cause of diarrheal illness in humans associated with antibiotic use and dysbiosis. We hypothesized that MET-1 would attenuate S. typhimurium infection through both antagonistic effects on the pathogen as well as effects on the host. However, we found that MET-1 had little effect on pathogen burden. Instead, MET-1 attenuated the systemic and localized effects of Salmonella infection and preserved intestinal tight junction protein expression.
MET-1 did not decrease S. typhimurium levels in the colon but systemic colonization was reduced in the infected animals treated with MET-1; levels of S. typhimurium in the small intestine were not measured, since MET-1 bacteria are derived from large intestine and are not expected to colonize the small intestine. However, the small intestine is a major point of Salmonella invasion and another study by Antunes et al. reported that fecal microbiota can inhibit the expression of invasion virulence factors by S. typhimurium26. In contrast to the latter report26, MET-1 did not have any effects on the ability of S. typhimurium to intracellularly invade intestinal epithelial cells when tested in an in vitro invasion assay. This may be due to the fact that the Lachnospiraceae species found by that study to be most inhibitory, such as C. citroniae and C. aldenense, are found within the C. clostridioforme subclade of the Lachnospiraceae family and this group is not represented in MET-1. Collectively, our results indicate that MET-1 confers protection against S. typhimurium primarily through its effects on multiple host functions, rather than through direct effects on Salmonella.
MET-1 was found to have only modest inhibition of Salmonella in time-kill assays and no effect in vivo. The interplay between MET-1, S. typhimurium and the host that occurs in the setting of the host inflammatory response to infection also likely contributes to this finding. Several studies have described the fitness advantage provided to facultative anaerobes in the environment of the inflamed intestine, replete with superoxide radicals, reactive oxygen species and reactive nitrogen species that may allow S. typhimurium to utilize alternate metabolic pathways and energy sources37,38. S. typhimurium has developed evolutionary strategies to utilize compounds reacting with organic sulfides and tertiary amines in the gut lumen as terminal electron acceptors for anaerobic respiration, thus allowing it to thrive in the gut under conditions of inflammation. Since MET-1 is composed primarily of anaerobic bacteria that cannot utilize these same metabolic pathways and carbon sources, we speculate that the inflamed milieu would provide favorable conditions for growth of S. typhimurium and may additionally contribute to the lack of MET-1 inhibition against S. typhimurium growth observed in vivo.
At the apical intestinal surface, TJ complexes between epithelial cells that involve cytoplasmic and membrane proteins such as claudin-1 and ZO-1 normally maintain the epithelial barrier39. By disrupting this and altering the distribution of TJ proteins40 S. typhimurium can increase paracellular permeability and membrane “leakiness” to provide an additional route of access to the basolateral compartment35,36. Our data indicate that MET-1-mediated preservation of tight junction protein expression is a probable mechanism for the decreased systemic effects of S. typhimurium. While upregulation of mucins such as MUC2 by probiotic commensal bacteria has been reported34,41, we found no evidence to support a MET-1 effect on intestinal mucus or MUC2. We chose to focus on MUC2, since this is the main mucin produced by goblet cells42 however goblet cells produce other mucins that could potentially be induced by exposure to MET-1 and the possibility exists that other mucins or other secretory products from goblet cells, induced by MET-1, may exist that could contribute to host protection against Salmonella.
There was a significant reduction in the serum cytokines involved in the maturation and recruitment of neutrophils (GM-CSF, MCP-1 eotaxin and MIP-1β) in infected animals that received MET-1. It is tempting to speculate that a MET-1-mediated decrease in the neutrophil population in the gut of infected MET-1 mice may have also contributed to decreased barrier disruption, since neutrophil transmigration across the epithelial barrier causes a breach in barrier function through release of neutrophil proteases such as elastase, which transiently degrade epithelial tight junctions and increase “leakiness” across the paracellular space43,44.
In assessing the microbiome of MET-1 treated and non-treated groups we found that some MET-1 isolates were identical to murine microbiota sequences that pre-existed in the mice (such as the Parabacteroides), whereas others (Acidaminococcus intestini, Roseburia faecalis/intestinalis, Bacteroides ovatus and Bifidobacterium adolescentis/longum) were unique to MET-1 and separable from what was originally in the mice at baseline, prior to receiving MET-1. However, limitations of the 16S rRNA gene profiling technique (which have been recognized in our previous human studies23) mean that it is impossible to be certain that changes in the murine gut microbiota brought about by MET-1 treatment were the result of colonization by MET-1 components; to effectively do this would require isolation of microbes from the murine gut, genome sequencing of these isolates and alignment of obtained sequences with MET-1 component genomes, which was beyond the scope of this work. We are currently working to identify unique sequence signatures in MET-1 genomes that can be used to differentiate MET-1 strains from others and which can be used in our future efforts to understand the nature of MET-1 component colonization in different hosts.
As a complex ecosystem, MET-1 has the potential to accomplish multiple core functions of the microbiota and to act through multiple mechanisms to protect the host. Here, a single dose of MET-1 resulted in improved outcomes - a potential advantage of using a defined, gut-derived microbial ecosystem acting through multiple mechanisms, over standard single-strain probiotics45,46. This study extends previous observations using defined microbial mixtures to treat recurrent C. difficile infection23,47,48. We demonstrate here that MET-1 confers protective properties in S. typhimurium infection both through attenuated inflammation and preservation of tight junction proteins, providing evidence that this defined microbial community may be useful for the treatment of other enteric infections in addition to C. difficile infection. Future studies of the complex interplay between host, microbiota and pathogen will be instrumental in leading to both a better understanding of gastrointestinal infectious disease and novel strategies of microbiota-based therapies.
Materials and Methods
Ethics statement
This study was carried out in accordance with the guidelines of the Canadian Council of Animal Care and was approved by Queen's University Animal Care Committee.
Bacterial culture
Salmonella enterica serovar Typhimurium SL1344 (Caliper Life Sciences, MA, USA), a streptomycin-resistant strain, was grown in Luria-Bertani (LB) broth (Bioshop Canada Inc., ON, Canada) supplemented with 100 μg/mL streptomycin at 37 oC for 18 hours as previously described49.
The derivation of MET-1 (Table 1), commensal colonic bacteria from a healthy human volunteer, is described elsewhere23. In brief, the 33 MET-1 isolates were cultured on fastidious anaerobe agar (FAA) (Lab M Ltd., Lancashire, UK) with or without 5% defibrinated sheep blood (Hemostat Laboratories, CA, USA) under anaerobic conditions. Plates were incubated at 37 °C for 2–3 days under strict anaerobic conditions in a Bugbox (Ruskinn, Maine, USA). Biomass was scraped directly into pre-reduced, filter-sterilized 0.9% saline using microbiological loops to achieve 3.5 × 109 CFU/mL23, previously determined to approximate the relative abundance of similar species in the stool samples of healthy North Americans (http://www.hmpdacc.org/).
Salmonella enterica serovar Typhimurium colitis model
The S. typhimurium model of colitis21 was initiated in 6–7 week old female C57BL/6 mice (Charles River, MA, USA). Mice were fasted for four hours prior to oral gavage with 20 mg of streptomycin (SteriMax, ON, Canada), after which mice had access to food and water ad libitum. Eighteen hours later mice were gavaged with either 100 μL of MET-1 (corresponding to 3.5 × 108 CFU) or vehicle control (0.9% saline). Four hours later mice were gavaged with 108 CFU of S. typhimurium or phosphate buffered saline (PBS). Weights were monitored daily and mice from each treatment group were euthanized 48 hours post-infection. Tissues and blood were collected for analysis as described below.
Bacterial enumeration in tissues and stool
Forty-eight hours post-infection spleens and colons were harvested for enumeration of S. typhimurium. Tissues were weighed, homogenized in 1 mL of sterile PBS and serial dilutions of homogenates plated on MacConkey agar containing 100 μg/mL streptomycin, incubated at 37 °C for 24 hours. Colonies were confirmed as S. typhimurium by PCR and an amplified band of the correct size on a 1% agarose gel was confirmed to be from S. typhimurium50. PCR primer sequences used50 were: forward primer 5′-AACAACGGCTCCGGTAATGA-3′ and reverse primer 5′-TGACAAACTCTTGATTCTGA-3′, targeting a putative cytoplasmic protein (310 bp product, STM4497) and forward primer 5′-TTTGGCGGCGCAGGCGATTC and reverse primer 5′ GCCTCCGCCTCATCAATCCG (423 bp product, STM3098), targeting a putative transcriptional regulator, both of which have been used to detect Salmonella species51. Cycling conditions: 95 °C for 3 min, (95 °C for 30 s, 58 °C for 30 s, 72 °C for 90 s) × 30; 72 °C for 5 min.
Multiplex bead assay to measure serum cytokine levels
Blood was collected via cardiac puncture 48 hours post-infection and a mouse cytokine magnetic bead kit (Bio-Plex Pro Mouse Cytokine multiplex, Bio-Rad, CA, USA) was used to measure serum IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-17A, eotaxin, G-CSF, GM-CSF, INF-γ, KC, MCP-1, MIP-1α, MIP-1β and TNF-α, as per the manufacturer’s instructions.
Immunohistochemistry
Immunofluorescence staining was carried out on formalin-fixed cecal tissues with the exception of MUC2, where tissues were fixed in Carnoy’s fixative52. Paraffin embedded tissues were sectioned (4 μm), hydrated and subjected to antigen retrieval in citric acid buffer (10 mM citric buffer, 2 mM EDTA, 0.05% Tween 20, pH 6.0).
For myeloperoxidase (MPO), claudin and NF-κB P65 staining, tissues were blocked with PBS containing 0.3 M glycine and 0.1% Tween 20, 3% bovine serum albumin (Bioshop Canada Inc.) and 10% goat serum (Sigma Aldrich), for one hour at room temperature. For MUC2, tissues were blocked with PBS with 5% fetal bovine serum (FBS) for 1 hour at room temperature. ZO-1 tissues were blocked with PBS-T containing 2% bovine serum albumin and 1% goat serum for 1 hour at room temperature.
Sections were stained with rabbit anti-MPO (1:100, Abcam, MA, USA) or rabbit anti-claudin-1 (1:200, Abcam), or rabbit anti-NF-κB P65 (1:50, Santa Cruz) or rabbit anti-MUC2 (1:100 dilution, Santa Cruz) or 1:100 mouse anti-ZO-1 (Invitrogen), in blocking buffer over night at 4 °C. Sections were washed with PBS-T three times, then incubated with Alexa 488-conjugated goat anti-rabbit immunoglobulin G (Invitrogen, CA, USA) at a 1:500 dilution (MPO, claudin, NF-κB and MUC2) or 1:100 (ZO-1) in blocking buffer. Tissues were incubated for one hour at room temperature for MPO, claudin, NF-κB, ZO-1 and two hours at room temperature for MUC2. Prolong antifade mounting media containing 4,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei (Invitrogen).
For MPO, claudin and NF-κB, images were obtained using an Olympus BX51 microscope and Image-Pro Plus software (Media Cybernetics, Inc., MD, USA). MUC2 images were visualized using an AxioImager M.1 microscope and AxioVision software (Carl Zeiss, Oberkochen, Germany). ZO-1 images were obtained using a Zeiss LSM 710 Laser Scanning confocal microscope.
Images used for quantification were taken at 400× magnification from at least two non-adjacent sections of cecum per mouse and 4 non-adjacent images were then used for each section to quantify. Images were taken from two independent experiments and each experiment had 3–4 mice per group. Total number of cells (determined by DAPI staining of nuclei) per view at 400× magnification was counted using ImagePro Plus software. An average of 700 cells (range: 540–1000 cells) was counted per view. The area of green fluorescence was measured using the ImagePro Plus software. The ratio of area of green versus total number of nuclei was used for the quantification53. Data were analyzed using a 1-way ANOVA with Bonferroni correction.
Alcian blue staining
Sections of mouse ceca were rehydrated, immersed in 3% acetic acid for 2 minutes and then stained with 1% (w/v) Alcian blue solution (8GX, Acros Organics, NJ, USA) in 3% acetic acid (pH 2.5) for 30 minutes. Tissues were rinsed in distilled water and counterstained with 0.1% nuclear fast red solution (Acros Organics) for 1 minute. All images were visualized using an Olympus BX60 microscope and Infinity Capture software (Lumenera Corporation, ON, Canada). Goblet cells were enumerated from stained tissues by counting goblet cells in 10 random high-powered fields/sample (400x magnification) spanning muscularis mucosa to surface epithelium.
Western blot analysis
Cecal tissues were sonicated in lysis buffer (10mM Tris, pH 7.6 containing 5 mM MgSO4, DNAse I, RNAse A and protease inhibitor cocktail; Roche, ON, Canada). Samples were incubated on ice for 20 minutes and centrifuged at 2,900 × g for 20 minutes at 4 °C. For fractionated samples, supernatant containing both cytosolic and membrane fractions were centrifuged at 21,000 × g for 45 minutes at 4 °C. The resulting supernatants comprised the soluble (S) cytosolic fractions. Pellets were resuspended in RIPA buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail), left on ice for 20 minutes and centrifuged at 15,000 × g for 20 minutes at 4 °C; the resulting supernatants comprised the insoluble (I) membrane fractions.
Samples of whole lysates, soluble fractions, or insoluble fractions were separated on a 12% SDS-PAGE gel, transferred overnight before blocking with 5% non-fat dry milk in TBST for 1 hour at room temperature. Membranes were then incubated with 1:1000 anti-ZO-1 (Life Technologies, ON, Canada), 1:1000 anti-claudin-1 (Life Technologies), or 1:10,000 anti-β-actin (Sigma Aldrich) overnight at 4 °C. After washing with TBS-T three times, horseradish peroxidase (HRP)-conjugated secondary antibodies were added (1:10,000 dilution, Cell Signaling, MA, USA) and membranes incubated for 1 hour at room temperature. After three more washes with TBS-T, PVDF membranes were exposed to chemiluminescence (ECL) reagents (Thermo Fisher Scientific, MA, USA) and exposed to autoradiographic film (VWR, ON, Canada) for band visualization. Protein levels were quantified by densitometry using ImageJ software.
Salmonella enterica serovar Typhimurium time-kill assay
MET-1 (initially 3.5 × 109 CFU/mL) was co-cultured with S. typhimurium (using different dilutions of MET-1, see below) and S. typhimurium viable counts monitored over time. Each well contained 107 CFU S. typhimurium in Wilkins Chalgren broth (Bioshop Canada Inc.) The overall bacterial CFU of MET-1: S. Tm. were as follows: MET-1 1:2 dilution = 3.5 × 109 CFU MET-1: S. typhimurium (350:1); 1:100 dilution = 7.0 × 107 CFU MET-1: S. typhimurium (7:1); 1:300 dilution = 2.3 × 107 CFU MET-1: S. typhimurium (2.3:1). Mixtures were incubated at 37 °C under anaerobic conditions (90% N2, 5% CO2 and 5% H2) for 24 hours and 100 μL aliquots withdrawn at 0, 1, 2, 3, 4, 5, 6, 7, 8, 12 and 24 hours, serial diluted and plated on MacConkey agar plates containing 100 μg/mL streptomycin. Plates were incubated at 37 °C for 24 hours and colonies were enumerated.
Salmonella enterica serovar Typhimurium invasion assay
The effect of MET-1 pretreatment on the susceptibility of Caco-2 cells (ATCC HTB-37) to S. typhimurium infection was assessed using an invasion assay based on a previous study54. Briefly, Caco-2 cells were seeded in 12-well plates and maintained in Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (37 °C in a 5% CO2, 95% air atmosphere). Wells containing 70–90% confluent monolayers54 were washed in DMEM and MET-1 or saline was added to each well (10% concentration). Cells were incubated at 37 °C for 1 hr, media were removed, cells were washed with PBS and fresh media were added. Washed live or heat-killed (boiled for 30 min) S. typhimurium (multiplicity of infection of 100:1) or saline (equivalent volume) were added to each well. Cells were incubated at 37 °C for 1 hr and washed with PBS. Non-internalized S. typhimurium cells were killed with gentamicin (100 μg/mL) for 1 hr at 37 °C. Media were removed, cells were washed with PBS and cells were treated with 0.1% Triton X-100 and incubated for 10 minutes. CFU/mL of S. typhimurium were calculated by serial dilutions plated onto MacConkey agar (Difco Laboratories, Becton, Dickinson and Company, MD, USA) supplemented with streptomycin (100 μg/mL).
Quantitative real-time PCR
An approximately 5 mm section of cecum tissue was immediately stored in RNAlater (Sigma Aldrich). Briefly, tissue was homogenized and RNA was extracted using an RNeasy Mini kit (Qiagen, ON, Canada) as per manufacturer’s instructions. RNA quality was assessed by absorbance at 260 nm (on a spectrophotometer) and by running on the Agilent 2100 Bioanalyzer (Agilent Technologies, ON, Canada). RNA was treated with RNAse free DNAse I (Invitrogen) to eliminate genomic DNA and quantitative RT-PCR (qRT-PCR) was performed in triplicates using the StepOnePlusTM Real-Time PCR System (Life Technologies). qRT-PCR was run using q-ScriptTM One-Step SYBR Green qRT-PCR kit (Quanta Biosciences, MD, USA) in a 10 μL reaction volume containing equal amounts of RNA (62 ng), 5 μL One-Step SYBR Green Master Mix, 0.2 μL qScript One-Step Reverse Transcriptase and gene specific forward (TNF-α, 200 nM; GAPDH 50 nM) and reverse (TNF-α, 50 nM; GAPDH, 150 nM) primers. Cycling conditions were as follows: 50 °C for 5 min, 95 °C for 2 min, (95 °C for 3 s, 60 °C for 30 s) × 40 cycles. The TNF-α and GAPDH primer sequences were as follows: TNF-α, forward 5′-CCACCACGCTCTTCTGTCTA-3′, reverse 5′-AGGGTCTGGGCCATAGAACT-3′; GAPDH, forward 5′-CGTCCCGTAGACAAAATGGT-3, reverse 5′-TTGATGGCAACAATCTCCAC-3′. The relative expression of TNF-α normalized to the expression of GAPDH (endogenous control) was calculated by the comparative CT (ΔΔCT) method55 using the vehicle control group as a reference group and one of the vehicle control group mice as a reference sample.
Statistical analysis
Results are expressed as the mean value with standard error of the mean (SEM). Statistical analyses were performed with GraphPad Prism version 5.0 (GraphPad Software, CA, USA). A two-tailed unpaired t-test, 2-way ANOVA with a Bonferroni correction, or 1-way ANOVA with Newman-Keuls or a Bonferroni correction was used where indicated. Statistical significance was set at a P value of <0.05.
Additional Information
How to cite this article: Martz, S.-L. E. et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci. Rep. 5, 16094; doi: 10.1038/srep16094 (2015).
References
Mueller, C. & Macpherson, A. J. Layers of mutualism with commensal bacteria protect us from intestinal inflammation. Gut 55, 276–284 (2006).
Bäckhed, F. et al. Defining a healthy human gut microbiome: current concepts, future directions and clinical applications. Cell Host Microbe 12, 611–622 (2012).
Sekirov, I., Russell, S. L., Antunes, L. C. M. & Finlay, B. B. Gut microbiota in health and disease. Physiol. Rev. 90, 859–904 (2010).
Endt, K. et al. The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Salmonella Diarrhea. PLoS Pathog 6, e1001097 (2010).
Chung, H. et al. Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell 149, 1578–1593 (2012).
Rajilić-Stojanović, M. et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141, 1792–1801 (2011).
Lee, K. N. & Lee, O. Y. Intestinal microbiota in pathophysiology and management of irritable bowel syndrome. World J. Gastroenterol. WJG 20, 8886–8897 (2014).
Hedin, C. R. et al. Altered intestinal microbiota and blood T cell phenotype are shared by patients with Crohn’s disease and their unaffected siblings. Gut 63, 1578–1586 (2014).
Willing, B. P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854.e1 (2010).
Kootte, R. S. et al. The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes. Metab. 14, 112–120 (2012).
Ley, R. E. Obesity and the human microbiome. Curr. Opin. Gastroenterol. 26, 5–11 (2010).
Brown, C. T. et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PloS One 6, e25792 (2011).
Larsen, N. et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS One 5, e9085 (2010).
Upadhyaya, S. & Banerjee, G. Type 2 diabetes and gut microbiome: at the intersection of known and unknown. Gut Microbes 6, 85–92 (2015).
Sekirov, I. et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76, 4726–4736 (2008).
Ubeda, C. et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120, 4332–4341 (2010).
Majowicz, S. E. et al., International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 50, 882–889 (2010).
Pavia, A. T. et al. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J. Infect. Dis. 161, 255–260 (1990).
Grisaru-Soen, G., Wysoki, M. G. & Keller, N. Risk factors for development of nontyphoid Salmonella bacteremia. Clin. Pediatr. (Phila.) 43, 825–829 (2004).
Nisbet, D. J. et al. Effect of a defined continuous-flow derived bacterial culture and dietary lactose on Salmonella typhimurium colonization in broiler chickens. Avian Dis. 37, 1017–1025 (1993).
Barthel, M. et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858 (2003).
Thiennimitr, P., Winter, S. E. & Bäumler, A. J. Salmonella, the host and its microbiota. Curr. Opin. Microbiol. 15, 108–114 (2012).
Petrof, E. O. et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome 1, 3 (2013).
Kamada, N., Chen, G. Y., Inohara, N. & Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690 (2013).
Valdez, Y., Ferreira, R. B. R. & Finlay, B. B. Molecular mechanisms of Salmonella virulence and host resistance. Curr. Top. Microbiol. Immunol. 337, 93–127 (2009).
Antunes, L. C. M. et al. Antivirulence activity of the human gut metabolome. mBio 5, e01183–01114 (2014).
Lara-Tejero, M. et al. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J. Exp. Med. 203, 1407–1412 (2006).
Fournier, B. M. & Parkos, C. A. The role of neutrophils during intestinal inflammation. Mucosal Immunol. 5, 354–366 (2012).
Arnold, J. W. et al. Tumor necrosis factor-alpha mediates the early pathology in Salmonella infection of the gastrointestinal tract. Microb. Pathog. 14, 217–227 (1993).
Yang, K. K. et al. Neutrophil influx in response to a peritoneal infection with Salmonella is delayed in lipopolysaccharide-binding protein or CD14-deficient mice. J. Immunol. Baltim. Md 1950 169, 4475–4480 (2002).
Cheminay, C., Chakravortty, D. & Hensel, M. Role of neutrophils in murine salmonellosis. Infect. Immun. 72, 468–477 (2004).
Strauss, R. R., Paul, B. B., Jacobs, A. A. & Sbarra, A. J. Mouse splenic peroxidase and its role in bactericidal activity. Infect. Immun. 5, 120–126 (1972).
Zarepour, M. et al. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect. Immun. 81, 3672–3683 (2013).
Bergstrom, K. S. B. et al. Muc2 Protects against Lethal Infectious Colitis by Disassociating Pathogenic and Commensal Bacteria from the Colonic Mucosa. PLoS Pathog 6, e1000902 (2010).
Liao, A. P. et al. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PloS One 3, e2369 (2008).
Köhler, H. et al. Salmonella enterica serovar Typhimurium regulates intercellular junction proteins and facilitates transepithelial neutrophil and bacterial passage. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G178–187 (2007).
Thiennimitr, P. et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. USA 108, 17480–17485 (2011).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010).
Denker, B. M. & Nigam, S. K. Molecular structure and assembly of the tight junction. Am. J. Physiol. 274, F1–9 (1998).
Boyle, E. C., Brown, N. F. & Finlay, B. B. Salmonella enterica serovar Typhimurium effectors SopB, SopE, SopE2 and SipA disrupt tight junction structure and function. Cell. Microbiol. 8, 1946–1957 (2006).
Tlaskalová-Hogenová, H. et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 8, 110–120 (2011).
Kim, Y. S. & Ho, S. B. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep. 12, 319–330 (2010).
Ginzberg, H. H. et al. Neutrophil-mediated epithelial injury during transmigration: role of elastase. Am. J. Physiol. Gastrointest. Liver Physiol. 281, G705–717 (2001).
Szabady, R. L. & McCormick, B. A. Control of neutrophil inflammation at mucosal surfaces by secreted epithelial products. Front. Immunol. 4, 220 (2013).
Symonds, E. L. et al. Bifidobacterium infantis 35624 protects against salmonella-induced reductions in digestive enzyme activity in mice by attenuation of the host inflammatory response. Clin. Transl. Gastroenterol. 3, e15 (2012).
Castillo, N. A., Perdigon, G. & de Moreno de LeBlanc, A. Oral administration of a probiotic Lactobacillus modulates cytokine production and TLR expression improving the immune response against Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol. 11, 177 (2011).
Lawley, T. D. et al. Targeted Restoration of the Intestinal Microbiota with a Simple, Defined Bacteriotherapy Resolves Relapsing Clostridium difficile Disease in Mice. PLoS Pathog 8, e1002995 (2012).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Zhang, Y. et al. Axin1 prevents Salmonella invasiveness and inflammatory response in intestinal epithelial cells. PloS One 7, e34942 (2012).
Kim, H.-J., Park, S.-H. & Kim, H.-Y. Comparison of Salmonella enterica serovar Typhimurium LT2 and non-LT2 salmonella genomic sequences and genotyping of salmonellae by using PCR. Appl. Environ. Microbiol. 72, 6142–6151 (2006).
McClelland, M. et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856 (2001).
Johansson, M. E. V. & Hansson, G. C. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue and localization of bacteria with FISH. Methods Mol. Biol. Clifton NJ 842, 229–235 (2012).
Lourenssen, S., Houpt, E. R., Chadee, K. & Blennerhassett, M. G. Entamoeba histolytica Infection and Secreted Proteins Proteolytically Damage Enteric Neurons. Infect. Immun. 78, 5332–5340 (2010).
Forbes, S. J., Eschmann, M. & Mantis, N. J. Inhibition of Salmonella enterica Serovar Typhimurium Motility and Entry into Epithelial Cells by a Protective Antilipopolysaccharide Monoclonal Immunoglobulin A Antibody. Infect. Immun. 76, 4137–4144 (2008).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Acknowledgements
This work was supported by the Crohn’s and Colitis Foundation of Canada (E.O.P).
Author information
Authors and Affiliations
Contributions
S.E.M. and J.A.K.M. designed and performed experiments, prepared figures, helped write the manuscript. Y.Z., C.N., S.H. and T.G. performed experiments, prepared figures. M.B., D.J.H and G.B.G performed data analysis. J.S., E.A.-V. and E.C.C. analyzed data, wrote sections of the manuscript. E.O.P. designed experiments, wrote the manuscript, provided oversight for the whole project. All authors discussed the results, reviewed the final manuscript.
Ethics declarations
Competing interests
E.O.P and E.A.V. are co-founders of Nubiyota and have filed a patent for MET-1 through Parteq Innovations (Queen’s University). The other authors have no competing interests to declare.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Martz, SL., McDonald, J., Sun, J. et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep 5, 16094 (2015). https://doi.org/10.1038/srep16094
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep16094
This article is cited by
-
Clostridium species as probiotics: potentials and challenges
Journal of Animal Science and Biotechnology (2020)
-
Antimicrobial Peptide JH-3 Effectively Kills Salmonella enterica Serovar Typhimurium Strain CVCC541 and Reduces Its Pathogenicity in Mice
Probiotics and Antimicrobial Proteins (2019)
-
Embracing the gut microbiota: the new frontier for inflammatory and infectious diseases
Clinical & Translational Immunology (2017)
-
A human gut ecosystem protects against C. difficile disease by targeting TcdA
Journal of Gastroenterology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.