Abstract
Aberrantly expressed long noncoding RNAs (lncRNAs) are associated with various cancers. However, the roles of lncRNAs in the pathogenesis of most cancers are unclear. Here, we report that the lncRNA FER1L4 (fer-1-like family member 4, pseudogene) acts as a competing endogenous RNA (ceRNA) to regulate the expression of PTEN (a well-known tumor suppressor gene) by taking up miR-106a-5p in gastric cancer. We observed that FER1L4 was downregulated in gastric cancer and that its level corresponded with that of PTEN mRNA. Both FER1L4 and PTEN mRNA were targets of miR-106a-5p. Further experiments demonstrated that FER1L4 downregulation liberates miR-106a-5p and decreases the abundances of PTEN mRNA and protein. More importantly, FER1L4 downregulation accelerated cell proliferation by promoting the G0/G1 to S phase transition. We conclude that one mechanism by which lncRNAs function in in tumorigenesis is as ceRNAs for tumor suppressor mRNAs.
Similar content being viewed by others
Introduction
Long noncoding RNAs (lncRNAs) regulate gene expression at many levels that include transcriptional, post-transcriptional and translational regulation1,2,3,4. Increasing numbers of studies have indicated that lncRNAs play key roles in tumorigenesis and may be used in the diagnosis of cancers3,5. Our previous study revealed that the fer-1-like family member 4, pseudogene (lncRNA-FER1L4) is strongly downregulated in gastric cancer6,7. However, the biological significance of this phenomenon is unknown.
Salmena and colleagues proposed a competing endogenous RNA (ceRNA) hypothesis to explain how different types of RNAs communicate with each other via microRNAs (miRNAs)8. According to this hypothesis, mRNAs, lncRNAs and other RNAs act as natural miRNA sponges to suppress intracellular miRNA function by using shared miRNA response elements (MREs)9,10,11. This hypothesis has been supported by numerous studies12,13,14,15. Because they are not actively translated, noncoding RNAs are thought to be highly effective ceRNAs8. Subsequent studies revealed that lncRNAs, including linc-MD115, lincRNA-RoR16,17, H1918, HOTAIR19,20, CARL21, lncRNA-ATB22 and lncRNA-BGL323, may function as ceRNAs.
Our previous work indicated that FER1L4 is a target of miR-106a-5p24. The well-known tumor suppressor gene-phosphatase and tensin homolog (PTEN) mRNA is also a validated target of miR-106a-5p12. In this study, we investigated whether FER1L4 can act as a ceRNA for PTEN mRNA through miR-106a-5p. We found that FER1L4 acted as a ceRNA to regulate PTEN expression by acting as a sponge for miR-106a-5p in gastric cancer. FER1L4 downregulation has been found to be a characteristic molecular change in gastric cancer6 and to lead to decreases in PTEN mRNA and protein levels. Because PTEN is a negative regulator of the cell cycle, we studied the effects of the downregulation of FER1L4 on the cell cycle; downregulation of FER1L4 by small interfering RNAs (siRNAs) increased cell proliferation by promoting the G0/G1 to S phase transition.
Results
FER1L4 and PTEN mRNA are downregulated in gastric cancer
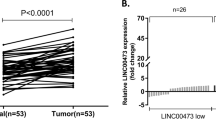
Our previous work revealed that the FER1L4 level is significantly lower in gastric cancer tissues than in paracancerous tissues6. In this study, we further found that FER1L4 levels in the human gastric cancer cell lines AGS, MGC-803 and SGC-7901 were lower than those in the human normal gastric epithelial cell line GES-1 (Fig. 1a). These results suggest that decreases in the level of FER1L4 may be associated with the occurrence of gastric cancer.
Expression of FER1L4 in gastric cancer cells and tissues.
(a) Expression of FER1L4 in a human normal gastric epithelial cell line and human gastric cancer cell lines. Data are presented as mean ± SD, n = 3. **P < 0.01, ***P < 0.001. Expression of FER1L4 (b) and PTEN (c) in the “FER1L4 high” and “FER1L4 low” subsets. n = 10, *P < 0.05.
Considering that FER1L4 and PTEN mRNA are targets of miR-106a-5p12,24 and that miR-106a-5p is highly expressed in gastric cancer25, we speculated that FER1L4 and PTEN mRNA may act as a pair of ceRNAs that are linked by miR-106a-5p. An important corollary of the ceRNA hypothesis is that ceRNAs coordinately regulate each other’s expression8. Based on the above findings, we investigated whether FER1L4 was coexpressed with PTEN in human samples. We measured FER1L4 and PTEN expression in 20 gastric cancer tissue samples by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The samples were sorted into two subsets (n = 10) according to the FER1L4 expression level (i.e., high and low FER1L4 groups, Fig. 1b). As shown in Fig. 1c, we observed significantly higher levels of PTEN mRNA in the high FER1L4 group than in the low FER1L4 group and vice versa. This coexpression is consistent with the ceRNA hypothesis.
FER1L4 and PTEN are targeted by miR-106a-5p in gastric cells
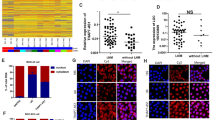
PTEN mRNA is one of the validated targets of miR-106a-5p12. In our previous study, the interaction between FER1L4 and miR-106a-5p was first predicted by miRcode and then confirmed by dual luciferase reporter assays24. However, we do not know whether miR-106a-5p effectively regulates FER1L4 and PTEN in gastric cells. To increase the miR-106a-5p level, we transfected miR-106a-5p mimics into the normal human gastric epithelial cell line GES-1 and the human gastric cancer cell lines, AGS, MGC-803 and SGC-7901. Next, we utilized qRT-PCR analysis to reveal that miR-106a-5p suppressed both FER1L4 and PTEN mRNA abundance in all of the tested gastric cell lines (Fig. 2a,b).
Moreover, to further test whether FER1L4 and PTEN expression levels were linked by miR-106a-5p, we decreased the miR-106a-5p level by transfection of its inhibitors into GES-1 and AGS cells. qRT-PCR analyses indicated that the transfection of miR-106a-5p inhibitors not only increased FER1L4 levels but also increased PTEN levels in both GES-1 and AGS cells (Fig. 2c). Because FER1L4 and PTEN mRNA are targets of miR-106a-5p, the knockdown of miR-106a-5p led to increases in free FER1L4 and PTEN mRNA.
Our previous study showed that by decreasing the expression of cyclin-dependent kinase (CDK) 1 and CDK2, the miR-106a inhibitors arrested gastric cancer cells at the G0/G1 and G2/M phases and then suppressed cell proliferation; and the results of animal experiments showed that the miR-106a inhibitors significantly suppressed tumor growth in a dose-dependent manner26. As a result, we assumed that the tumor suppression effect of miR-106a inhibitors might indirectly through the overexpression of FER1L4.
Effects of FER1L4 downregulation on miR-106a-5p and PTEN expression
The ceRNA hypothesis proposes that the downregulation of miRNA targets will result in freeing of the same miRNA molecules8. For example, GAS5-siRNA significantly reduces the endogenous GAS5 level while simultaneously increasing the miR-21 level27. Thus, we sought to determine whether the downregulation of FER1L4 would influence miR-106a-5p and the levels of its targets. In our previous study, we designed a siRNA against FER1L4 and effectively reduced FER1L4 levels in the normal human gastric epithelial cell line GES-1 and the human gastric cancer cell lines AGS, MGC-803 and SGC-790124. Here, we transfected FER1L4-siRNA into GES-1, AGS, MGC-803 and SGC-7901 cells and qRT-PCR analyses revealed that the miR-106a-5p levels in the gastric cells were increased by the knockdown of FER1L4 (Fig. 3).
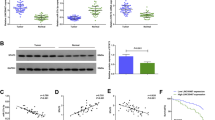
If FER1L4 functions as a ceRNA, its downregulation might free additional miR-106a-5p. This miR-106a-5p would target PTEN mRNA and trigger the downregulation of PTEN. We knocked down FER1L4 in GES-1, AGS, MGC-803 and SGC-7901 cells and monitored PTEN expression by qRT-PCR and Western blot. FER1L4 knockdown resulted in decreased levels of PTEN mRNA and protein (Fig. 4).
Expression of PTEN in a human normal gastric epithelial cell line and human gastric cancer cell lines after FER1L4 knockdown.
(a) PTEN mRNA levels detected by qRT-PCR. Data are presented as mean ± SD, n = 3. NC, negative control. *P < 0.05, **P < 0.01. (b) Representative cropped results of Western blot analyses. (c) Results of Western blot analyses of three independent experiments. NC, negative control. **P < 0.01, ***P < 0.001. The gels were run under the same experimental conditions. The blots were processed in parallel.
FER1L4 regulates the cell cycle and cell proliferation
PTEN acts as a negative regulator of the cell cycle by suppressing the phosphoinositide 3-kinase (PI3K)-AKT pathway28. Because perturbations of the FER1L4 level markedly affected PTEN expression, we decided to investigate the effects of disrupting FER1L4 on the cell cycle and cell proliferation. Flow cytometry demonstrated that FER1L4 downregulation promoted the G0/G1 to S phase transition in GES-1, AGS, MGC-803 and SGC-7901 cells (Fig. 5). Moreover, FER1L4 knockdown also accelerated cell proliferation in all of the gastric cell lines (Fig. 6).
Discussion
Studies have revealed that ceRNAs play important roles in post-transcriptional regulation and are involved in oncogenesis and cancer progression12,13,17,19,20,23,29,30,31,32,33. Our previous study found that several lncRNAs may be associated with gastric cancer via their actions as ceRNAs24. In the present study, we focused on FER1L4 and PTEN mRNA because both of these RNAs are targets of miR-106a-5p, a typical onco-miRNA12,24,25. FER1L4 was downregulated in gastric cancer tissues6 and gastric cancer cells (Fig. 1a). FER1L4 was coexpressed with PTEN such that the upregulation of FER1L4 led to greater expression of PTEN and vice versa (Fig. 1b,c). To test whether FER1L4 downregulation resulted in a reduction in the level of PTEN expression by freeing miR-106a-5p, we knocked down FER1L4. This knockdown resulted in an increase in miR-106a-5p (Fig. 3) and a decrease in PTEN mRNA and protein levels (Fig. 4). Furthermore, FER1L4 downregulation accelerated cell proliferation (Fig. 6) by promoting the G0/G1 to S phase transition (Fig. 5). These results indicate that FER1L4 and PTEN mRNA are a pair of ceRNAs that are linked by miR-106a-5p.
Indeed, the FER1L4-PTEN interaction may be associated with additional miRNAs. FER1L4 and PTEN share 33 types of MREs as predicted by miRcode34. It has been found that several transcripts, such as PTENP129,35, CNOT6L12, VAPA12, VCAN30, ZEB213 and lncRNA-BGL323, act as ceRNAs of PTEN mRNA that are mediated by pools of miRNAs (Fig. 7). These transcripts share different types of MREs with PTEN mRNA and are connected in a complex ceRNA network. Furthermore, these transcripts may also be ceRNAs of other RNAs with which they share different MREs. For example, VCAN acts as a ceRNA in the regulation of not only PTEN but also RB1 and CD3430,36. ceRNAs may contain various MREs and each miRNA may target hundreds of transcripts. Different pathways may interact with each other via ceRNA cross-talk14. In addition, whether ceRNAs can alter miRNA function in vivo requires further investigation37,38.
The study of ceRNA interactions represents a new approach to examining complex post-transcriptional regulatory networks. Cancer is caused by alterations in various pathways. We could design specific ceRNAs to alter these pathways for therapeutic purposes.
Recently, circular RNAs (circRNAs) were included as new members of ceRNAs39,40,41,42,43. A circRNA named CDR1as that contains approximately 70 MREs functions as a ceRNA that sequesters miR-7 away from its targets44,45. The ceRNA world is becoming increasingly more complex and fascinating. To better understand ceRNAs systematically, several groups have developed in silico mathematical ceRNA models to describe the characteristics of ceRNAs and ceRNA networks46,47,48,49,50. Moreover, several algorithms and databases, such as Linc2GO51, starBase52, lnCeDB53 and Cupid54, have been developed to predict ceRNA interactions. These tools facilitate ceRNA studies.
ceRNAs can not only be used to explain biological phenomena, such as autophagy, apoptosis and morphogenesis55,56,57, but can also be developed as miRNA inhibitors, such as short tandem target mimic (STTM)58 and circular miRNA sponges59. For example, circular miRNA sponges have been found to exhibit excellent anticancer effects59. These findings indicate that ceRNAs might represent new therapeutic approaches to cancer and other diseases60. Further investigations may help us to understand how ceRNAs contribute to oncogenesis and tumor metastasis. ceRNA disturbances may be associated with many diseases61,62,63,64.
In conclusion, we found that FER1L4 was minimally expressed in gastric cancer. Via its functions as a ceRNA, FER1L4 liberated miR-106a-5p, downregulated PTEN expression and affected cell growth.
Methods
The methods were carried out in accordance with the approved guidelines.
All experimental protocols were approved by The Human Research Ethics Committee from Ningbo University.
Tissue samples
Biopsy samples of gastric cancer tissues were obtained at the Yinzhou Hospital Affiliated to Ningbo University School of Medicine from patients who had not undergone previous radiotherapy or chemotherapy. The Human Research Ethics Committee of Ningbo University approved all aspects of the protocols. Written informed consent was obtained from all of the subjects.
Cells and culture conditions
The human gastric epithelial cell line GES-1 was obtained from the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences (Beijing, China). The human gastric cancer cell lines, AGS, MGC-803 and SGC-7901 were obtained from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). All cell lines were grown in RPMI Medium 1640 (Life Technologies, Carlsbad, CA, USA) plus 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere with 5% CO2. The cells were counted using a TC10 Automated Cell Counter (Bio-Rad, Hercules, CA, USA).
Transient transfection
For the transfection of the miRNA mimics and siRNAs, GES-1, AGS, MGC-803 and SGC-7901 cells (2 × 105) were seeded in 6-well plates. The following day, they were transfected with 120 nM of miRNA mimic or siRNA using Lipofectamine 2000 Reagent (Life Technologies). The sequence of the miR-106a-5p mimic was 5′-AAAAGUGCUUACAGUGCAGGUAG-3′. The sequence of the miR-106a-5p inhibitor was 5′-CUACCUGCACUGUAAGCACUUUU-3′. The sequence of the negative control was 5′-CAGUACUUUUGUGUAGUACAA-3′. The sequence of the FER1L4 siRNA was 5′-CAGGACAGCUUCGAGUUAATT-3′ (sense) and 5′-UUAACUCGAAGCUGUCCUGTT-3′ (antisense). The sequences of the negative control siRNAs were 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense). These sequences were synthesized by GenePharma Co., Ltd. (Shanghai, China).
RNA extraction
Total RNA was extracted using TRIzol Reagent (Life Technologies) according to the manufacturer’s protocol. The RNA quantity was measured with a SmartSpec Plus spectrophotometer (Bio-Rad). The RNA purity was evaluated according to the A260/A280 ratio.
qRT-PCR analysis
qRT-PCRs of the lncRNAs and mRNAs were performed using a GoTaq 2-Step RT-qPCR System (Promega, Madison, WI, USA) in a Mx3005P QPCR System (Stratagene, La Jolla, CA, USA) according to the manufacturer’s protocol. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. The FER1L4 primers were as follows: forward, 5′-CCGTGTTGAGGTGCTGTTC-3′; reverse, 5′-GGCAAGTCCACTGTCAGATG-3′. The PTEN primers were as follows: forward, 5′-GTTTACCGGCAGCATCAAAT-3′; reverse, 5′-CCCCCACTTTAGTGCACAGT-3′. The GAPDH primers were as follows: forward, 5′-AAGGTGAAGGTCGGAGTCAA-3′; reverse, 5′-AATGAAGGGGTCATTGATGG-3′.
qRT-PCR of the mature miRNAs was performed using miScript II RT Kits (Qiagen, Hilden, Germany), miScript SYBR Green PCR Kits (Qiagen) and miScript Primer Assays (miR-106a-5p primer and RNU6-2 primer; Qiagen) in the Mx3005P QPCR System (Stratagene) according to the manufacturer’s protocol. RNU6-2 was used as a control.
All experiments were performed in triplicate. Relative quantification of gene expression was performed by the 2−ΔΔCt method65,66.
Western blot
The cells were collected and lysed with cell lysis buffer for Western blotting (Beyotime, Haimen, China). The proteins (30 μg per lane) were separated on 12% SDS-polyacrylamide gels and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Immunoblotting of the membranes was performed using the following primary antibodies: anti-PTEN (CST, Danvers, MA, USA) and anti-β-actin (4A Biotech, Beijing, China). The signals were revealed after incubation with the recommended secondary antibodies using an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE, USA). β-actin was used as the control.
Cell cycle analysis
The cells were washed in PBS and fixed in 75% ice-cold ethanol at − 20 °C overnight. After rehydrating with ice-cold PBS, the cells were stained with PI/RNase Staining Buffer (BD Biosciences, San Jose, CA, USA) and analyzed by flow cytometry on a FACSCalibur Flow Cytometer (BD Biosciences) using CellQuest Pro software.
Cell proliferation assays
The proliferation assays were performed in E-Plate 96 using a Real-Time Cell Analyzer (RTCA) (ACEA Biosciences, San Diego, CA, USA) according to the manufacturer’s protocol.
Statistical analysis
The data are presented as the means ± the SDs. The differences between groups were evaluated with two-tailed Student’s t-tests using SPSS Statistics 20.0 software (IBM, Armonk, NY, USA). P < 0.05 was considered to be statistically significant.
Additional Information
How to cite this article: Xia, T. et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci. Rep. 5, 13445; doi: 10.1038/srep13445 (2015).
Change history
16 September 2015
The HTML version of this Article previously published noted an incorrect current address for Tian Xia. This has now been corrected in the HTML; the PDF version of the paper was correct from the time of publication.
References
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81, 145–166 (2012).
Batista, P. J. & Chang, H. Y. Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307 (2013).
Xu, C. et al. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol 35, 9701–976 (2014).
Geisler, S. & Coller, J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14, 699–712 (2013).
Chen, S., Li, P., Xiao, B. & Guo, J. Long noncoding RNA HMlincRNA717 and AC130710 have been officially named as gastric cancer associated transcript 2 (GACAT2) and GACAT3, respectively. Tumour Biol 35, 8351–8352 (2014).
Liu, Z. et al. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol 35, 9613–9617 (2014).
Song, H. et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med 11, 225 (2013).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011).
Ebert, M. S., Neilson, J. R. & Sharp, P. A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4, 721–726 (2007).
Ebert, M. S. & Sharp, P. A. Emerging roles for natural microRNA sponges. Curr Biol 20, R858–R861 (2010).
Bak, R. O. & Mikkelsen, J. G. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA 5, 317–333 (2014).
Tay, Y. et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357 (2011).
Karreth, F. A. et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395 (2011).
Sumazin, P. et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147, 370–381 (2011).
Cesana, M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011).
Wang, Y. et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog and Sox2 in human embryonic stem cell self-renewal. Dev Cell 25, 69–80 (2013).
Hou, P. et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 5, e1287 (2014).
Kallen, A. N. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 52, 101–112 (2013).
Liu, X. H. et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 13, 92 (2014).
Ma, M. Z. et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer 13, 156 (2014).
Wang, K. et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 5, 3596 (2014).
Yuan, J. H. et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 (2014).
Guo, G. et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 34, 1768–1779 (2015).
Xia, T. et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep 4, 6088 (2014).
Xiao, B. et al. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta 400, 97–102 (2009).
Liao, Z. W., Deng, H. X., Zhang, G. P., Zhou, H. & Guo, J. M. Effect of oncogenic microRNA-106a on growth of normal gastric mucous epithelial cells and gastric cancer cells. Chin J Pathophysiol 27, 1885–1889 (2011).
Zhang, Z. et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ 20, 1558–1568 (2013).
Song, M. S., Salmena, L. & Pandolfi, P. P. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 13, 283–296 (2012).
Poliseno, L. et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038 (2010).
Lee, D. Y. et al. Expression of versican 3′-untranslated region modulates endogenous microRNA functions. PLoS One 5, e13599 (2010).
Wang, L. et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis 34, 1773–1781 (2013).
Kumar, M. S. et al. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 505, 212–217 (2014).
Rutnam, Z. J., Du, W. W., Yang, W., Yang, X. & Yang, B. B. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat Commun 5, 2914 (2014).
Jeggari, A., Marks, D. S. & Larsson, E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 28, 2062–2063 (2012).
Johnsson, P. et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 20, 440–446 (2013).
Fang, L. et al. Versican 3′-untranslated region (3′-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J 27, 907–919 (2013).
Denzler, R., Agarwal, V., Stefano, J., Bartel, D. P. & Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 54, 766–776 (2014).
Bosson, A. D., Zamudio, J. R. & Sharp, P. A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol Cell 56, 347–359 (2014).
Jeck, W. R. et al. Circular RNAs are abundant, conserved and associated with ALU repeats. RNA 19, 141–157 (2013).
Taulli, R., Loretelli, C. & Pandolfi, P. P. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol 20, 541–543 (2013).
Jeck, W. R. & Sharpless, N. E. Detecting and characterizing circular RNAs. Nat Biotechnol 32, 453–461 (2014).
Starke, S. et al. Exon circularization requires canonical splice signals. Cell Rep 10, 103–111 (2015).
Li, P. et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 444, 132–136 (2015).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Ala, U. et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci USA 110, 7154–7159 (2013).
Bosia, C., Pagnani, A. & Zecchina, R. Modelling competing endogenous RNA networks. PLoS One 8, e66609 (2013).
Figliuzzi, M., Marinari, E. & De Martino, A. MicroRNAs as a selective channel of communication between competing RNAs: a steady-state theory. Biophys J 104, 1203–1213 (2013).
Nitzan, M., Steiman-Shimony, A., Altuvia, Y., Biham, O. & Margalit, H. Interactions between distant ceRNAs in regulatory networks. Biophys J 106, 2254–2266 (2014).
Figliuzzi, M., De Martino, A. & Marinari, E. RNA-based regulation: dynamics and response to perturbations of competing RNAs. Biophys J 107, 1011–1022 (2014).
Liu, K., Yan, Z., Li, Y. & Sun, Z. Linc2GO: a human LincRNA function annotation resource based on ceRNA hypothesis. Bioinformatics 29, 2221–2222 (2013).
Li, J. H., Liu, S., Zhou, H., Qu, L. H. & Yang, J. H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42, D92–D97 (2014).
Das, S., Ghosal, S., Sen, R. & Chakrabarti, J. lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS One 9, e98965 (2014).
Chiu, H. S. et al. Cupid: simultaneous reconstruction of microRNA-target and ceRNA networks. Genome Res 25, 257–267 (2015).
Inui, M., Montagner, M. & Piccolo, S. miRNAs and morphogen gradients. Curr Opin Cell Biol 24, 194–201 (2012).
Xu, J., Wang, Y., Tan, X. & Jing, H. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 8, 873–882 (2012).
Ge, D. et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy 10, 957–971 (2014).
Tang, G. et al. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58, 118–125 (2012).
Liu, Y. et al. Construction of circular miRNA sponges targeting miR-21 or miR-221 and demonstration of their excellent anticancer effects on malignant melanoma cells. Int J Biochem Cell Biol 45, 2643–2650 (2013).
Giza, D. E., Vasilescu, C. & Calin, G. A. MicroRNAs and ceRNAs: therapeutic implications of RNA networks. Expert Opin Biol Ther 14, 1285–1293 (2014).
Hansen, T. B., Kjems, J. & Damgaard, C. K. Circular RNA and miR-7 in cancer. Cancer Res 73, 5609–5612 (2013).
Karreth, F. A. & Pandolfi, P. P. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov 3, 1113–1121 (2013).
Tay, Y., Rinn, J. & Pandolfi, P. P. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014).
Karreth, F. A., Tay, Y. & Pandolfi, P. Target competition: transcription factors enter the limelight. Genome Biol 15, 114 (2014).
Yu, X. et al. Growth inhibitory effects of three miR-129 family members on gastric cancer. Gene 532, 87–93 (2013).
Shao, Y. et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 120, 3320–3328 (2014).
Acknowledgements
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (Grant No. LY14C060003), the Applied Research Project on Nonprofit Technology of Zhejiang Province (Grant No. 2014C33222), the National Natural Science Foundation of China (Grant No. 81171660) and the K.C. Wong Magna Fund of Ningbo University.
Author information
Authors and Affiliations
Contributions
T.X. and J.G. conceived and designed the experiments. T.X., S.C., Z. J., Y.S., X.J. and P.L. performed the experiments. T.X., B.X. and J.G. analyzed the data. T.X. and J.G. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Xia, T., Chen, S., Jiang, Z. et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep 5, 13445 (2015). https://doi.org/10.1038/srep13445
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep13445
This article is cited by
-
Pseudogenes and the associated ceRNA network as potential prognostic biomarkers for colorectal cancer
Scientific Reports (2022)
-
Cre/Lox-based RMCE for Site-specific Integration in CHO Cells
Biotechnology and Bioprocess Engineering (2021)
-
Long Noncoding RNAs in Gastrointestinal Cancer: Tumor Suppression Versus Tumor Promotion
Digestive Diseases and Sciences (2021)
-
Correlation between expression levels of lncRNA FER1L4 and RB1 in patients with colorectal cancer
Molecular Biology Reports (2021)
-
Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance
Molecular Cancer (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.