Abstract
RE1-Silencing Transcription factor (REST) has a well-established role in regulating transcription of genes important for neuronal development. Its role in cancer, though significant, is less well understood. We show that REST downregulation in weakly invasive MCF-7 breast cancer cells converts them to a more invasive phenotype, while REST overexpression in highly invasive MDA-MB-231 cells suppresses invasiveness. Surprisingly, the mechanism responsible for these phenotypic changes does not depend directly on the transcriptional function of REST protein. Instead, it is driven by previously unstudied mid-size (30–200 nt) non-coding RNAs (ncRNAs) derived from the first exon of an alternatively spliced REST transcript: REST-003. We show that processing of REST-003 into ncRNAs is controlled by an uncharacterized serine/arginine repeat-related protein, SRRM3. SRRM3 expression may be under REST-mediated transcriptional control, as it increases following REST downregulation. The SRRM3-dependent regulation of REST-003 processing into ncRNAs has many similarities to recently described promoter-associated small RNA-like processes. Targeting ncRNAs that control invasiveness could lead to new therapeutic approaches to limit breast cancer metastasis.
Similar content being viewed by others
Introduction
One of the most important events in the natural history of breast cancer is the transformation of intraductal carcinoma to invasive disease1. If left untreated, invasive cells can progress to widespread metastases associated with a worsening prognosis. A major goal of cancer research is to identify the underlying mechanisms that regulate the in situ to invasive transition, in order to support new therapeutic strategies that would benefit cancer patients.
Recent work establishes a strong correlation between cancer invasiveness and the loss of RE1-Silencing Transcription factor (REST), a well-characterized protein best known for suppressing neuronal genes during development2,3,4,5,6,7. REST activity is suppressed during neurogenesis partly by reduced expression6,7 as well as by ubiquitin-induced proteolytic degradation8. Suppression or loss of REST function may also result from alternative splicing. An alternatively spliced form of REST, called REST4, has been identified in several cancer types that do not express normal REST transcripts (e.g., neuroblastoma9, small cell lung cancer5,10 and breast cancer4). In neurons, alternative splicing of REST into REST4 is controlled by a neuronal-specific Ser/Arg repeat protein (nSR100/SRRM4). Interestingly, REST can directly silence SRRM4 expression, thereby preventing alternative splicing and thus acting as an on/off switch for neuronal gene expression11,12. A similar REST-dependent alternative splicing mechanism could conceivably play a role in acquisition of neuronal-like properties of breast cancer cells, enabling them to become invasive13.
In addition to REST4 splicing, extensive alternative splicing of REST pre-mRNA has been detected in cancer tissues and cell lines14. At least 45 variants have been identified, some of which may code for modified proteins and others that are non-coding RNAs (ncRNAs). Significant generation of short ncRNAs in proximity to transcriptional start sites (TSSs) from human cells has recently been described15. An intriguing example is the promoter-associated short non-coding RNAs (PASRs) that appear to be derived from the 5’ ends of known protein-coding genes15,16,17,18. PASRs are generally in the size class of less than 200 nt but longer than typical inhibitory RNAs (i.e., >30 nt)19.
Here, we used RNA deep sequencing to identify a previously uncharacterized alternatively spliced form of the REST gene, REST-003, that plays a key role in controlling cancer cell invasiveness. REST-003 is a non-coding RNA that is processed into a complicated series of sense and antisense ncRNAs with sizes ranging from 30–200 nt. The alternative splicing of REST transcript in invasive cells appears to be controlled by a previously unstudied SR protein, SRRM3, as well as by the REST protein itself. REST expression results in downregulation of both SRRM3 and REST-003 transcript. REST-003 levels correlate positively with cancer cell invasiveness and knockdown of REST-003 is sufficient to convert invasive cancer cells to a non-invasive phenotype. Our results reveal regulatory interactions among REST, REST-003 and a cancer cell-specific splicing activator (SRRM3) that may coordinate gene regulation required for development of the invasive cancer phenotype in breast cancer.
Results
REST levels correlate negatively with cancer invasion in breast cancer cell lines
We explored the REST-dependent invasive phenotype in more detail using two breast cancer cell lines: MDA-MB-231, which is strongly invasive and MCF-7, which is weakly invasive. We first confirmed the invasive potential of each cell line using Matrigel invasion chamber assays20 (Fig. S1A). In agreement with published studies2,3, REST mRNA and protein levels were higher in MCF-7 relative to MDA-MB-231 cells (Fig. S1B, C).
We next downregulated REST in MCF-7 cells using two small interfering RNAs (siRNAs) (si-REST_1 and si-REST_2; see Fig. S1D and Table S1). Treated cells exhibited increased invasiveness in Matrigel assays (Fig. S1E). We then overexpressed wild-type (wt) REST21 in MDA-MB-231 cells by transfection with REST cDNA and observed decreased Matrigel invasion (Fig. S1F). These data confirm a negative correlation between REST expression and invasiveness2,4.
An alternatively spliced transcript of REST, REST-003, positively correlates with invasiveness
To identify candidate genes that mediate invasiveness, we performed RNA-sequencing (RNA-seq) analysis (GEO accession# GSE63610) of MCF-7 and MDA-MB-231 cells. RNA-seq data suggested downregulation of REST transcript levels in MCF-7 cells following treatment with 2 different siRNAs designed to target the protein coding region of REST. REST expression also increased when REST cDNA was transfected into MDA-MB-231 cells (Fig. 1A). These changes were confirmed by measuring REST expression using qRT-PCR in multiple biological replicates (Fig. 2A,B; Fig. S1D). REST transcript levels did not differ significantly between the two cell lines (Fig. 1D,E).
Altering REST-003 ncRNA expression in si-REST-treated MCF-7 and REST-overexpressed MDA-MB-231 cells.
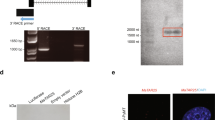
REST expression was calculated as the number of reads normalized to the median number of reads for all genes in each sample. (A) Expression of REST in siRNA-treated MCF-7 and REST-overexpressed MDA-MB-231 cells. (B) Relative expression of each REST splice transcript was normalized to total REST expression and transcript length in siRNA-treated MCF-7 and REST-overexpressed MDA-MB-231 cells. Some exons in the gene model are shared between multiple transcripts (Fig. S2A). For these, relative expression was calculated as the percentage of REST-aligned reads assigned to a specific transcript/transcript-group normalized by the mappable length of the transcript or transcript-group. (C) Schematic diagram of the REST gene and its splice variant transcripts, including illustrations of annotated REST exons and locations of primers employed for the identification of REST splice variants. The constitutive transcript (REST-001) and alternative spliced variants are shown in red, REST-002 in green, REST-003 in yellow and REST-004 in blue. REST-001 and/or REST-002 can produce the REST protein (wt-REST) that has the complete coding region containing 9 zinc fingers with DNA binding activity (purple) and two repressor domains (pink) necessary for recognizing the RE1 elements and exhibiting repressor function. Forward and reverse primers are indicated by right (numbers) and left (letters) arrows, respectively. (D) (E) Detection (D) and expression levels (E) of ncRNA REST-003 (3-B primer pair) and coding REST RNA (R-N or R-M primer pair) in MCF-7 and MDA-MB-231 cells by qRT-PCR. (D) Following PCR amplification, samples were loaded on 4% agarose gel with a 100-bp marker. (E) Expression levels were normalized to GAPDH, CyclophilinA, and/or Actin, converted to MNE and presented as Relative Expression (Rel. Exp.) after normalization to control samples. Biological replicates are shown on the bar graph as mean plus SEM (paired t-test).
Effect of REST on REST-003 ncRNA expression in cells and effect of si-REST-003 on MDA-MB-231 invasiveness.
(A) (B) Effect of REST on expression of REST-003 ncRNAs in si-REST-treated MCF-7 (A) and REST-overexpressed MDA-MB-231 cells (B) by qRT-PCR. MCF-7 cells treated with a non-REST siRNA (si-GAPDH) (A) and MDA-MB-231 cells transfected with EGFP or mt-REST cDNA6 (lacking two repressor domains) (B) served as controls. ‘REST Rel. Exp.’ refers to REST‐001 (primer set R‐N). (C) Detection of REST-003 ncRNA by qRT-PCR (left) and invasiveness by a Matrigel invasion chamber (right) after treating MDA-MB-231 cells with si-REST-003. (D) Reduced REST-003 expression by si-REST-003 treatment in MDA-MB-231 cells using qRT-PCR. More than 50% of REST-003 transcripts were reduced by si-REST-003 relative to si-C (scramble). REST transcript expression [REST-001 (R-001), R-M primer pair] was not changed by si-REST-003 (si-R-003) treatment. For qRT-PCR data, expression levels were normalized to GAPDH, CyclophilinA and/or Actin, converted to MNE and presented as Rel. Exp. after normalization to control cells (A, B) or si-C samples (D). Biological replicates are shown on the bar graph as mean plus SEM [one-way ANOVA with Friedman test for multiple comparisons for (A, B) and paired t-test for (D)].
We estimated the relative expression of each alternatively spliced form of REST based on the EMBL gene model using our RNA-seq pipeline22,23 (Fig. 1B). We found an alternatively spliced product (ASP) of REST, REST-003, whose expression was low in MCF-7 and high in MDA-MB-231 cells (Fig. 1B, Fig. S2A). Due to the complex nature of REST alternative splicing in cancer cells14 as well as the small size of RNA-seq reads, we performed additional experiments to confirm this result and determine its role in invasiveness.
Only four REST ASPs are catalogued in the Ensembl Human Genome Browser database (version 75; http://uswest.ensembl.org/index.html). We confined our analysis to these forms (REST-001, REST-002, REST-003 and REST-004), which are illustrated schematically in Fig. 1C along with the REST gene (also see Fig. S2B and Supplementary Information). A translation initiation codon is present in Exon 2 (E2). REST-001 and REST-002 produce full-length REST protein but contain different 5’ untranslated regions (UTRs). REST-004 lacks the E2 initiation codon and may thus produce non-coding RNA (ncRNA). REST-003 contains the initiation codon but lacks other parts of the E2 coding sequence. Since no available data identify REST-003 as a protein-coding gene, we analyzed the structures of the REST ASPs with qRT-PCR, using specific primers to distinguish the presence or absence of the E2 initiation codon (Fig. 1C). Primers flanking the E2 initiation codon or the middle part of the coding region (R-N and R-M primer pairs) detected high REST expression in MCF-7 cells and low expression in MDA-MB-231 cells (Fig. 1D,E). We obtained a similar result with primers flanking the E2 initiation codon but confined to sequences present only in REST-003 (5-A primer pair; Fig. S3A, B). When testing primers that exclude the initiation codon but are specific to REST-003 (3-B primer pair), we observed low expression in MCF-7 cells (Fig. 1D,E) and other weakly invasive breast cancer cell lines (Fig. S4A, B) and high expression in MDA-MB-231 cells (Fig. 1D,E, Fig. S4A, B). Similarly, the invasive bladder cancer cell line, T24/83, expressed REST-003 at higher levels than the non-invasive RT112/84 line (Fig. S4C, D). These results support our RNA-seq data and suggest that only ncRNA derived from REST-003 correlates positively with invasiveness.
REST-003 expression is negatively controlled by REST
We next established the effect of REST modulation on REST-003 expression using RNA-seq and qRT-PCR. Both methods indicated enhanced REST-003 expression (>2 fold) following REST downregulation in MCF-7 cells (Fig. 1B, Fig. 2A). Expression of the potential coding RNAs (REST-002, primer pair 2-A or REST-003, primer pair 5-A) decreased relative to control expression (Fig. S3A), a pattern similar to that of REST-001 (Fig. 2A and Fig. S3A). Conversely, overexpression of REST in MDA-MB-231 cells resulted in decreased REST-003 expression (Fig. 1B, Fig. 2B and Fig. S3B). These results suggest that increased expression of REST-003 ncRNAs (following loss of REST) may mediate breast cancer cell invasiveness.
REST-003 plays an important role in regulating invasiveness
To verify the role of REST-003 in controlling invasiveness, we knocked down its expression in MDA-MB-231 cells with siRNA (si-REST-003). The si-REST-003-treated MDA-MB-231 cells exhibited decreased REST-003 expression (>50%) and reduced Matrigel invasion (>50%) relative to control cells treated with scrambled RNA (si-C; Fig. 2C,D). Treated cells did not show a change in REST-001 expression (Fig. 2C,D). This implicates a primary role for REST-003 in regulating invasiveness that is, at least in part, independent of REST protein expression.
REST-003 expression is controlled by an uncharacterized serine/arginine repeat-related protein, SRRM3, whose expression may be under REST-mediated transcriptional control
We next questioned how REST downregulation could result in increased REST-003 expression. In neuronal cells, REST transcript is alternatively spliced to produce a REST4 protein, which activates gene expression by competing with REST for RE-1 DNA binding sites24. This alternative splicing is mediated by a neural-specific serine/arginine (SR) repetitive matrix 4 protein, SRRM4 (also known as nSR100)25. We did not detect any SRRM4 expression by RNA-seq following REST overexpression in MDA-MB-231 cells or in MCF-7 cells treated with si-REST_2 (Fig. 3A). There was, however, a significant change in expression of a related gene, SRRM3 (Fig. 3A), which has no previously documented function. SRRM3 expression increases >2 fold in MCF-7 cells following treatment with si-REST RNAs, though its expression in MCF-7 is much higher than that in MDA-MB-231, as estimated by our standard pipeline analysis22,23 (Fig. 3A). We validated this increase using qRT-PCR (Fig. S3C and Table S1). In contrast, SRRM3 expression decreased by ~3 fold (CuffDiff and our pipeline) in MDA-MB-231 cells following REST overexpression (Fig. 3A). We confirmed these data with qRT-PCR (Fig. S3D).
Effect of REST on SRRM3 expression in cells and effect of SRRM3 on REST-003 ncRNA expression and invasiveness in MDA-MB-231 cells.
(A) Expression of different SRRM subfamilies in siRNA-treated MCF-7 and REST-overexpressed MDA-MB-231 cells by our pipeline analysis22,23 of RNA-seq. (B) N-terminal sequences of SRRM3, cfw21 (S. cerevisiae) and SRRM2 (H. sapiens) were compared using the ClustalW2 program. Identical residues in cwf21 domains are in red font for all three SR-related proteins. Stars indicate identical residues in SRRM3 and SRRM2. (C) Effect of si-SRRM3 on REST-003 ncRNA expression in MDA-MB-231 cells using qRT-PCR. (D) (E) Effect of different siRNA treatments on MDA-MB-231 Matrigel invasiveness (D) and REST-003 ncRNA expression (E). Expression levels were normalized to GAPDH, CyclophilinA and/or Actin, converted to MNE and presented as Rel. Exp. after normalization to control cells. Biological replicates are shown on the bar graph as mean plus SEM [paired t-test for (C) and one-way ANOVA with Dunnet test for multiple comparisons for (E)].
SRRM3 contains a cwf21 domain, suggesting interaction with SR proteins26. It also contains SR-rich domains scattered throughout its sequence (Fig. 3B). However, it lacks a canonical RNA recognition motif (RRM) thought to be necessary for alternative splicing. SRRM3 may thus be an “SR-related protein”27 that enhances transcription not by splicing, but in a manner similar to the RSR-2 protein in C. elegans28. We hypothesized that increased REST-003 expression and invasiveness were mediated by increased levels of SRRM3 following REST downregulation. To support this notion, when SRRM3 expression is suppressed in MDA-MB-231 cells using siRNA (si-SRRM3), we observed lower SRRM3 and REST-003 expression (Fig. 3C). Importantly, SRRM3 suppression also reduced MDA-MB-231 Matrigel invasiveness (Fig. 3D). Co-transfection of MDA-MB-231 cells with si-REST-003 and si-REST_2 eliminated the change in REST-003 expression as well as the reduction in invasiveness (Fig. 3D,E), suggesting that SRRM3 regulatory control of REST-003 is positioned downstream of REST protein.
REST-003 is processed into mid-size non-coding RNAs as both sense and anti-sense sequences
REST-003 appears to be expressed as a ~150-nt-long mid-size non-coding RNA (ncRNA) positioned within the first exon of REST mRNA (Fig. 1B,C). Recent findings reveal many new small- and mid-size ncRNAs that are enriched at the 5’ boundaries of some human genes15,16. We investigated the potential presence of a cluster of REST-003 ncRNAs using northern blot analysis of the 5’ region of REST (E1-3 region; Fig. 4A) and found several ncRNAs derived from this region (Fig. S5). Sequences with a length of ~70–150 nt were especially enriched in MDA-MB-231 cells and include both sense (S) and anti-sense (AS) sequences (Fig. 4B,C), an expression pattern similar to that of PASRs that are not yet functionally defined15,16. The ~75- and ~87-nt-long REST-003 S and the ~75-, ~87- and ~135-nt-long REST-003 AS ncRNA sequences were most highly expressed in MDA-MB-231 relative to MCF-7 cells. Since REST modulation affects REST-003 expression (Fig. 2A,B), we performed northern analysis in MDA-MB-231cells overexpressing REST and in MCF-7 cells treated with si-REST_2. We found expression of the ~75- and ~87-nt REST-003 S and the ~135-nt REST-003 AS ncRNAs to be negatively regulated by REST (Fig. 4B,C, blue boxes).
Expression pattern of REST-003 and its downregulating effect on MDA-MB-231 cells.
(A) Schematic picture of ncRNAs and coding RNAs transcribed from the E1-3 region. Many new ncRNAs that are enriched at the 5’ boundary of the REST gene (E1-3) are predicted as S (yellow) and AS (purple) sequences. (B) (C) Differential expression of REST-003 in MCF-7 and MDA-MB-231 cells using northern blot analysis. RNA samples were prepared from each cell line transfected with controls and different siRNAs, as indicated. Hybridizations were performed using32 P-labeled DNA oligonucleotide probes complementary to the S and AS transcripts probes: AS (B*) sequence of E1-3 for S (B) and S (3*) sequence for AS (C) detection. Human U6 RNA (~105 nt) was probed as an internal control. (D) Genes downregulated by REST-003 downregulation using RNA-seq analysis. Downregulated gene expression in si-REST-003-treated cells is shown using DESeq from our pipeline (adjusted P-value < 0.05; yellow). The downregulated genes were compared with published RNA-seq data35 from MCF-7 and MDA-MB-231 cell lines (BCCLs, purple), 42 triple negative (TNBC) tissues and 58 non-malignant control tissues (blue). (E) Schematic of the regulatory interactions among REST, REST-003 and SRRM3 that may coordinate gene regulation required for development of the invasive phenotype.
We also investigated the levels of the ncRNAs following treatment of MDA-MB-231 cells with si-SRRM3. While expression of the ~75- and 135-nt REST-003 AS ncRNAs were downregulated, REST-003 S ncRNA was unaffected (Fig. 4B,C, green boxes). Additionally, at least five larger (>200 nt) REST-003 S and AS bands were highly expressed in MCF-7 cells, similar to REST-001 (Fig. S5). Notably, we did not detect any ~21–23 nt double-stranded RNAs potentially derived from Dicer processing of REST-003. Taken together, our data indicate at least eight RNA variants derived from REST primary transcript (Fig. S5). Three appear to be ncRNAs that likely promote invasiveness (Fig. 4B,C). The other five could potentially code for modified proteins and/or produce other ncRNAs that inhibit invasiveness, similar to REST protein.
REST-003 mid-size non-coding RNAs are involved with immune, defense, wounding and inflammatory responses, as well as cancer cell invasion and/or extravasation for metastasis
We next sought a link between REST-003 expression, REST target genes and pathways related to invasion by performing RNA-seq analysis (GEO accession# GSE63610) of MDA-MB-231cells treated with si-REST-003 (targeting the E1-3 region). Fifty-six genes from DESeq were differentially expressed in the treated versus control samples (Fig. 4D, Fig. S6A). Ten were downregulated in the treated sample, while the other 46 were upregulated. Six of the downregulated genes (PLEC, SYK, STK35, SLC35B2, CUL4a and EPCAM) are known to facilitate cancer cell invasion and/or extravasation for metastasis29,30,31,32,33,34 (Fig. 4D). We compared these genes with published RNA-seq data from breast cancer cell lines and tissues35. Five genes (PLEC, ANXA10, EHF, SLC4A, CUL4A) were expressed highly in MDA-MB-231 relative to MCF-7 cells and three (MAGED1, SYK, EPCAM) were expressed more in triple-negative tissues relative to non-invasive control tissues (Fig. 4D). We classified the 46 upregulated genes in the treated sample with DAVID functional analysis (Fig. S6A, B) and found more than 20% to be related to immune, defense, wounding and inflammatory responses (Fig. S6C). Interestingly, neither REST nor its canonical neuronal target genes was affected by knockdown of REST-003 ncRNAs. These results indicate that REST-003 ncRNAs play an important role in cancer cell invasion that is largely independent of REST and REST target gene function.
Discussion
Many classes of ncRNAs associated with the transcriptional start sites (TSSs) of genes have been described18. Some examples include PASRs, TSS-associated RNAs (TSSa-RNAs), promoter upstream transcripts (PROMPTs) and transcription initiation RNAs (tiRNAs). Though the biological functions of these classes are poorly defined, most are believed to be involved in transcriptional regulation. In this study, we identified a new class of mid-size ncRNAs derived from a specific alternative splice form of the REST gene (REST-003). We propose that these ncRNAs provide a key regulatory function by controlling expression of genes important for cancer cell invasiveness. They may thus represent a new functional example of a PASR-like process (Fig. 4E).
One key technical aspect of our approach was to perform RNA deep sequencing without first selecting polyadenylated RNA. This allowed identification of transcripts derived from the first exon of an alternatively spliced REST transcript (REST-003), which are apparently further processed into mid-size (30–200 nt) ncRNAs. As expected from alternative splicing, REST-003 levels correlate negatively with those of REST-001 (the protein coding form of the REST gene). More importantly, we found that REST-003 levels correlate positively with cancer cell invasiveness in several cancer cell lines.
Subtypes of breast cancer are often classified with markers such as estrogen receptor (ER), progesterone receptor (PR) and HER2 that relate to clinical outcomes. At least four subtypes are recognized based on different expression patterns for these markers36,37,38. Ranked from most invasive to least invasive, these include36,39: basal-like triple negative (ER−, PR−, HER2−) = HER2+ > luminal B (ER+, PR+/−, HER2+) > luminal A (ER+, PR+/−, HER2−). We found that this order correlates well with levels of REST-003 (Fig. S4A, B). Low REST-003 levels were observed in MCF-7, T47D (luminal A), BT474 (luminal B), SKBR3 (HER2+) and MDA-MB-468 (basal-like) cell lines, all of which are non-invasive in a Matrigel assay (Fig. S4A, B). Conversely, high REST-003 levels were observed in basal-like triple negative and invasive MDA-MB-231 cells (Fig. 1D,E; Fig. S4A, B). Similarly, an invasive bladder cancer cell line (T24/83) exhibited high levels of REST-003 relative to non-invasive RT112/84 bladder cancer cells (Fig. S4C, D). These results suggest that REST-003 could be used as a prognostic marker to access invasive potential of breast and other types of cancer cells. Further studies will be required to establish this more firmly.
Our data suggest a possible mechanism for regulating alternative splicing of the REST primary transcript into REST-001 (or alternative protein coding forms) and REST-003. We found that REST itself negatively regulates expression of SRRM3, which suggests a positive association between the expression of SRRM3 and REST‐003 (Fig. 3A,C). SRRM3 has not yet been functionally characterized. We detected increased SRRM3 expression following downregulation of REST in MCF-7 cells and decreased expression following overexpression of REST in MDA-MB-231 cells (Fig. 3A), suggesting direct or indirect control of REST alternative splicing in addition to autoregulation via REST.
The alternative splicing networks in neuronal cells and cancer cells are clearly different. Neuronal cells express SRRM4, a known splicing regulator of REST25, while breast cancer cells lack SRRM4 expression (Fig. 3A) (with the exception of small cell lung cancer5). As a consequence, splicing in invasive MDA-MB-231 cells is not regulated by SRRM4. We propose that REST-003 expression is rather controlled by SRRM3. Regulatory networks for alternative splicing are complex and are still not fully defined40. Additional work is required to directly demonstrate this proposed mechanism favoring invasiveness and REST-003 in cancer cells.
Our findings show that REST downregulation results in increased production of REST-003, which is processed into many different sizes of ncRNAs (Fig. 4B,C). At least 3 of these processed ncRNAs are more prevalent (75- and 85-nt S and 135-nt AS) (Fig. 4B,C). Conversely, overexpression of REST results in a decrease in these same ncRNA fragments (Fig. 4B,C). These data suggest that further characterization focusing on these specific REST-003-derived ncRNAs may be the most significant factor controlling invasiveness. Interestingly, only reduction of the 135-nt AS product is observed following downregulation of SRRM3 (Fig. 4C, green boxes). Perhaps other splicing factors are involved in regulation, or other processing pathways remain to be discovered (Fig. 4E). Further studies could establish whether they act together or separately in regulating invasiveness as well as whether they are important in other cancer types.
Following si-REST-003 treatment of MDA-MB-231 cells, an impressive subset of invasive-related genes was downregulated (PLEC, SYK, STK35, SLC35B2, CUL4A and EPCAM). These genes are known to be important not only for cell line invasiveness (Fig. 4D), but also for cancer tumor progression to metastasis29,30,31,32,33,34. This suggests that REST-003 ultimately plays an important role in controlling invasiveness through a gene expression program. Other RNA-seq data from breast cancer cell lines and tissues35 provide supporting information for this hypothesis. For example, PLEC, ANXA10, EHF, SLC4A and CUL4A are expressed at high levels in MDA-MB-231 relative to MCF-7 cells and MAGED1, SYK and EPCAM are expressed at higher levels in triple-negative cancer tissues relative to non-invasive control tissues (Fig. 4D). In addition, REST-003 appears to be involved not only in regulating invasiveness, but also in immune, defense, wounding and inflammatory responses by upregulating expression of genes in these pathways (Fig. S6). Each of these genes may thus be regulated in different ways (i.e., increased or decreased) by the REST-003 mid-size ncRNAs. Our RNA-seq data place the focus on REST-003-derived mid-size ncRNAs.
In conclusion, a mechanistic role for REST-003 and SRRM3 in regulating cancer cell invasiveness suggests novel therapeutic approaches for limiting breast (and other types of) cancer invasion and metastasis.
Methods
Cell culture, transfection, qRT-PCR and northern blot
MDA-MB-231 and MCF-7 cancer cell lines were obtained and cultured as described previously41. Lipofectamine 2000 (Invitrogen) was used for all transfection experiments unless otherwise specified. For siRNA transfection, we used DharmaFect (Thermo Scientific) or RNAiMax (Invitrogen) according to the manufacturer’s instructions. siRNA sequences are provided in Supplementary Table 1.
For qRT-PCR, cDNAs were made from the total RNAs treated with DNase as described in our previous study42. Gene-specific q-PCR primer or probe sets (Table S1) for human genes, GAPDH, CyclophilinA and Actin and equivalent amounts of cDNA generated as a template were used for qRT-PCR. Reactions were performed for each sample using SSoFast EvaGreen Supermix (Bio-Rad) or TaqMan Universal PCR Master Mix (Invitrogen) with a CFX-96 system (Bio-Rad). For each sample, expression of marker genes was normalized to GAPDH, CyclophilinA or Actin [mean normalized expression (MNE)]. MNEs were normalized to control samples to present relative expression (Rel. Exp.). Northern blots were performed as described previously43.
Matrigel invasion assay
Invasion was measured using BD BioCoat Matrigel Invasion Chambers (BD Biosciences) according to the manufacturer’s instructions, as described in our previous study41.
Library preparation, sequencing and analysis of RNA-seq
Ribosomal RNA (rRNA) was depleted from 100 ng of total RNA using a Ribo-Zero Magnetic Gold kit (Epicentre) according to the manufacturer’s protocol. Libraries from rRNA-depleted samples were prepared using a TruSeq RNA Sample Preparation kit v2 (Illumina) following the recommended protocol starting from the RNA fragmentation stage. Purification of polyadenylated RNA was omitted. Libraries were pooled (4 samples per pool), clustered on cBOT (Illumina) and sequenced on HiSeq2000, each pool in one lane. We performed single-end 100 bp reads. Reads were mapped using TopHat followed by data analysis using Cufflinks and Cuffdiff software, as well as our own pipeline22,23.
For tissue data, expression reads of downregulated genes by si-REST-003 treatment were averaged from TNBC and controls by our pipeline22,23 assay of RNA-seq data35 (Table S2) and compared to each other.
Statistical analysis
Bar graphs show the mean ± SEM of biological replicates (n). Detailed statistical methods are indicated in the figure legends.
Additional Information
How to cite this article: Lee, N.S. et al. Non-coding RNAs derived from an alternatively spliced REST transcript (REST-003) regulate breast cancer invasiveness. Sci. Rep. 5, 11207; doi: 10.1038/srep11207 (2015).
References
Cowell, C. F. et al. Progression from ductal carcinoma in situ to invasive breast cancer: revisited. Molecular oncology 7, 859–869, doi:10.1016/j.molonc.2013.07.005 (2013).
Reddy, B. Y., Greco, S. J., Patel, P. S., Trzaska, K. A. & Rameshwar, P. RE-1-silencing transcription factor shows tumor-suppressor functions and negatively regulates the oncogenic TAC1 in breast cancer cells. Proc Natl Acad Sci USA 106, 4408–4413, doi:10.1073/pnas.0809130106 (2009).
Lv, H. et al. Expression and functions of the repressor element 1 (RE-1)-silencing transcription factor (REST) in breast cancer. Journal of cellular biochemistry 110, 968–974, doi:10.1002/jcb.22610 (2010).
Wagoner, M. P. et al. The transcription factor REST is lost in aggressive breast cancer. PLoS genetics 6, e1000979, doi:10.1371/journal.pgen.1000979 (2010).
Shimojo, M., Shudo, Y., Ikeda, M., Kobashi, T. & Ito, S. Small cell lung cancer-specific isoform of RE1-silencing transcription factor (REST) is regulated by neural-specific Ser/Arg repeat-related protein of 100 kDa (nSR100). Molecular cancer research : MCR, doi:10.1158/1541-7786.MCR-13-0269 (2013).
Chong, J. A. et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957 (1995).
Schoenherr, C. J. & Anderson, D. J. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363 (1995).
Qureshi, I. A. & Mehler, M. F. Regulation of non-coding RNA networks in the nervous system--what’s the REST of the story? Neuroscience letters 466, 73–80, doi:10.1016/j.neulet.2009.07.093 (2009).
Palm, K., Metsis, M. & Timmusk, T. Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain research. Molecular brain research 72, 30–39 (1999).
Coulson, J. M., Edgson, J. L., Woll, P. J. & Quinn, J. P. A splice variant of the neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: a potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer Res 60, 1840–1844 (2000).
Zheng, S. & Black, D. L. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends in genetics : TIG, doi:10.1016/j.tig.2013.04.003 (2013).
Zheng, S. et al. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nature neuroscience 15, 381–388, S381, doi:10.1038/nn.3026 (2012).
Neman, J. et al. Human breast cancer metastases to the brain display GABAergic properties in the neural niche. Proc Natl Acad Sci USA 111, 984–989, doi:10.1073/pnas.1322098111 (2014).
Chen, G. L. & Miller, G. M. Extensive alternative splicing of the repressor element silencing transcription factor linked to cancer. PLoS One 8, e62217, doi:10.1371/journal.pone.0062217 (2013).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108, doi:10.1038/nature11233 (2012).
Kapranov, P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488, doi:10.1126/science.1138341 (2007).
Morris, K. V. & Mattick, J. S. The rise of regulatory RNA. Nature reviews. Genetics 15, 423–437, doi:10.1038/nrg3722 (2014).
Zovoilis, A. et al. The expression level of small non-coding RNAs derived from the first exon of protein-coding genes is predictive of cancer status. EMBO reports 15, 402–410, doi:10.1002/embr.201337950 (2014).
Esteller, M. Non-coding RNAs in human disease. Nature reviews. Genetics 12, 861–874, doi:10.1038/nrg3074 (2011).
Albini, A. et al. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer research 47, 3239–3245 (1987).
Tapia-Ramirez, J., Eggen, B. J., Peral-Rubio, M. J., Toledo-Aral, J. J. & Mandel, G. A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc Natl Acad Sci U S A 94, 1177–1182 (1997).
Souaiaia, T., Frazier, Z. & Chen, T. ComB: SNP calling and mapping analysis for color and nucleotide space platforms. Journal of computational biology : a journal of computational molecular cell biology 18, 795–807, doi:10.1089/cmb.2011.0027 (2011).
Wang, Y. et al. RseqFlow: workflows for RNA-Seq data analysis. Bioinformatics 27, 2598–2600, doi:10.1093/bioinformatics/btr441 (2011).
Shimojo, M., Paquette, A. J., Anderson, D. J. & Hersh, L. B. Protein kinase A regulates cholinergic gene expression in PC12 cells: REST4 silences the silencing activity of neuron-restrictive silencer factor/REST. Molecular and cellular biology 19, 6788–6795 (1999).
Raj, B. et al. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Molecular cell 43, 843–850, doi:10.1016/j.molcel.2011.08.014 (2011).
Grainger, R. J., Barrass, J. D., Jacquier, A., Rain, J. C. & Beggs, J. D. Physical and genetic interactions of yeast Cwc21p, an ortholog of human SRm300/SRRM2, suggest a role at the catalytic center of the spliceosome. Rna 15, 2161–2173, doi:10.1261/rna.1908309 (2009).
Leoutsakou, T., Talieri, M. & Scorilas, A. Prognostic significance of the expression of SR-A1, encoding a novel SR-related CTD-associated factor, in breast cancer. Biological chemistry 387, 1613–1618, doi:10.1515/BC.2006.201 (2006).
Fontrodona, L. et al. RSR-2, the Caenorhabditis elegans ortholog of human spliceosomal component SRm300/SRRM2, regulates development by influencing the transcriptional machinery. PLoS genetics 9, e1003543, doi:10.1371/journal.pgen.1003543 (2013).
Banyard, J. et al. Identification of genes regulating migration and invasion using a new model of metastatic prostate cancer. BMC cancer 14, 387, doi:10.1186/1471-2407-14-387 (2014).
Saucedo-Cuevas, L. P. et al. CUL4A contributes to the biology of basal-like breast tumors through modulation of cell growth and antitumor immune response. Oncotarget 5, 2330–2343 (2014).
Goyal, P., Behring, A., Kumar, A. & Siess, W. STK35L1 associates with nuclear actin and regulates cell cycle and migration of endothelial cells. PLoS One 6, e16249, doi:10.1371/journal.pone.0016249 (2011).
Kamiyama, S. et al. Expression and the role of 3’-phosphoadenosine 5’-phosphosulfate transporters in human colorectal carcinoma. Glycobiology 21, 235–246, doi:10.1093/glycob/cwq154 (2011).
Krisenko, M. O., Cartagena, A., Raman, A. & Geahlen, R. L. Nanomechanical Property Maps of Breast Cancer Cells As Determined by Multiharmonic Atomic Force Microscopy Reveal Syk-Dependent Changes in Microtubule Stability Mediated by MAP1B. Biochemistry, doi:10.1021/bi500325n (2014).
Sugiyama, N. et al. In vivo selection of high-metastatic subline of bladder cancer cell and its characterization. Oncology research 20, 289–295 (2013).
Varley, K. E. et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat 146, 287–297, doi:10.1007/s10549-014-3019-2 (2014).
Vargo-Gogola, T. & Rosen, J. M. Modelling breast cancer: one size does not fit all. Nature reviews. Cancer 7, 659–672, doi:10.1038/nrc2193 (2007).
Kao, J. et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One 4, e6146, doi:10.1371/journal.pone.0006146 (2009).
Neve, R. M. et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer cell 10, 515–527, doi:10.1016/j.ccr.2006.10.008 (2006).
Sorlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98, 10869–10874, doi:10.1073/pnas.191367098 (2001).
Calarco, J. A., Zhen, M. & Blencowe, B. J. Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. Rna 17, 775–791, doi:10.1261/rna.2603911 (2011).
Hwang, J. Y. et al. Investigating contactless high frequency ultrasound microbeam stimulation for determination of invasion potential of breast cancer cells. Biotechnology and bioengineering, doi:10.1002/bit.24923 (2013).
Lee, N. S. et al. A novel dual-color reporter for identifying insulin-producing beta-cells and classifying heterogeneity of insulinoma cell lines. PLoS One 7, e35521, doi:10.1371/journal.pone.0035521 (2012).
Lee, N. S. et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nature biotechnology 20, 500–505, doi:10.1038/nbt0502-500 (2002).
Acknowledgements
We thank Derek Sieburth (USC) for discussions and critical reading of the manuscript, Jessica Alluin and John J. Rossi (City of Hope) for northern hybridization help and Gail Mandel (Vollum Institute, Oregon Health and Science University) for providing wt- and mt-REST cDNA constructs. This work was supported by the National Institutes of Health grant P41-EB2182 (K.K.S.) and a USC Zumberge grant (R.H.C. and K.K.S.).
Author information
Authors and Affiliations
Contributions
N.S.L. conceived and designed experiments. N.S.L. and P.M.S. wrote the manuscript. N.S.L. executed experiments constructing plasmids with A.B., J.Y.L., M.S. and all other molecular and cell biological assays with A.B. N.S.L., A.B. and Y.N. performed Matrigel invasion assays. N.S.L. and A.B. prepared RNA samples and carried out qRT-PCR. O.V.E. and J.H (CuffDiff), T.S. (our pipeline), J. K and K.W. (DESeq & DAVID) and J.A.K. made all RNA-seq libraries and performed bioinformatic analyses. A.C.W. helped write the manuscript. P.M.S., A.C.W., M.F.P., K.W., O.V.E. contributed helpful discussions/advice. H-J.L., K.K.S. and R.H.C. edited the manuscript with helpful suggestions. All authors reviewed the manuscript.
RNA-seq generated during this study has been deposited in GEO under accession number GSE63610. The authors declare no competing financial interest. A provisional patent pertaining to the results presented in the paper was filed.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lee, N., Evgrafov, O., Souaiaia, T. et al. Non-coding RNAs derived from an alternatively spliced REST transcript (REST-003) regulate breast cancer invasiveness. Sci Rep 5, 11207 (2015). https://doi.org/10.1038/srep11207
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep11207
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.