Abstract
Infection with high-risk human papillomaviruses (HPVs) causes cervical cancer. E6 oncoprotein, an HPV gene product, inactivates the major gatekeeper p53. In contrast, its isoform, TAp73β, has become increasingly important, as it is resistant to E6. However, the intracellular signaling mechanisms that account for TAp73β tumor suppressor activity in cervix are poorly understood. Here, we identified that IER3 is a novel target gene of TAp73β. In particular, TAp73β exclusively transactivated IER3 in cervical cancer cells, whereas p53 and TAp63 failed to do. IER3 efficiently induced apoptosis and its knockdown promoted survival of HeLa cells. In addition, TAp73β-induced cell death, but not p53-induced cell death, was inhibited upon IER3 silencing. Moreover, etoposide, a DNA-damaging chemotherapeutics, upregulated TAp73β and IER3 in a c-Abl tyrosine kinase-dependent manner and the etoposide chemosensitivity of HeLa cells was largely determined by TAp73β-induced IER3. Of interest, cervical carcinomas from patients express no observable levels of two proteins. Thus, our findings suggest that IER3 is a putative tumor suppressor in the cervix and the c-Ab1/p73β/IER3 axis is a novel and crucial signaling pathway that confers etoposide chemosensitivity. Therefore, TAp73β and IER3 induction would be a valuable checkpoint for successful therapeutic intervention of cervical carcinoma patients.
Similar content being viewed by others
Introduction
Cervical cancer is one of the most common cancers and the second leading cause of cancer-related death in women worldwide1. More than 99% of cervical cancer develops upon infection with human papilloma viruses (HPVs). Among over 120 HPVs, 15 of them are thought to cause cervical cancer, with HPV 16 and 18 being the two major types that account for more than 70% of all cases2,3. Viral E6 and E7 are two critical oncoproteins responsible for cervical cancer development from high-risk HPVs and dysregulate cell proliferation, apoptosis and genome instability4.
TP73 (p73) and p63 are homologues of the tumor suppressor p53 and they exhibit overlapping and unique roles5. Although p53 is the major cellular “gatekeeper” that inhibits tumor development, p53 is not functional in most of cervical cancers because E6 oncoprotein prevents p53 function by targeting p53 for degradation6,7. Unlike with p53, HPV E6 protein does not physically interact with p738 and the ectopic expression of TAp73 isoform efficiently inhibits the growth of E6-expressing HPV-positive cervical cancer cells9,10,11. Two isoforms of p73, α and β, contain transactivation (TA) domains required for the transcriptional regulation of their target genes, which induce apoptosis and cell cycle arrest5.
Immediate early response gene 3 (IER3), also known as IEX-1, Dif-2, gly96, or p22/PRG-1, is an early response gene rapidly induced by a wide range of stimuli, including growth factors, cytokines, DNA-damage and viral infection12. IER3 is an evolutionally conserved gene present in diverse species, including Caenorhabditis elegans and Drosophila melanogaster. In the human genome, IER3 does not have other close homologues and its sequence is extremely well conserved among mammals. IER3 mRNA is widely expressed in most human tissues, with more abundant expression in epithelial tissues with high cell turnover13. Its transcription is regulated by p53, Sp1, c-Myc and NF-κB in a synergistic or opposing manner in different cells14,15,16. IER3 regulates multiple cellular processes, including apoptosis17,18,19,20, proliferation21,22, differentiation21,23 and DNA repair24 and its response varies depending on cellular context. Although the mechanism underlying the IER3-mediated induction of apoptosis is not clearly understood at present, its involvement with the BCL-2 family, which is the pivotal regulator of cell survival and death, has been demonstrated; the presence of functional BIM is required for IER3-induced apoptosis. IER3 interacts with MCL-1 and IER3 inhibits the expression of BCL-2 and BCL-xL, respectively17,25.
In the present study, we identified IER3 as a specific transcriptional target gene of TAp73, but not p53, in cervical cancer cells. In addition, we demonstrated that IER3 is a critical mediator of TAp73β-induced cell death in cervical carcinoma cells and etoposide chemosensitivity of HeLa cells was largely governed by TAp73β-induced IER3. Furthermore, we found that IER3 and TAp73β expression levels were undetectable in cervical carcinoma tumors, implying that downregulation of these two proteins could be implicated in the development of cervical cancer.
Results
IER3 is a specific transcriptional target gene of TAp73β in cervical cancer cells
To investigate transcriptional activities of the p53 family proteins p53, p63 and p73 on IER3, we generated a human IER3 promoter construct (-1384 bp) possessing a previously known p53-binding element16 and performed luciferase reporter assays in different cell lines. Overexpressed TAp73β specifically activated IER3 gene transcription in a dose-dependent manner in human cervical cancer cells, including the HeLa, KB, Caski and SiHa cell lines, which express E6 or E7 oncoproteins from high-risk HPV types 18 or 16, whereas neither p53 nor TAp63 were able to stimulate IER3 promoter activation (Figure 1A). Similar results were confirmed at the mRNA level of IER3 as determined by a real-time PCRs analysis (Supplementary Figure 1A). In contrast, we did not observe this specific regulation in other cell lines, including human embryo kidney (293T), colorectal carcinoma (HCT116 and SW480) and ovarian adenocarcinoma (SK-OV-3) cells (Figure 1A and Supplementary Figure S1B). In addition, knockdown of TAp73β by small-interfering RNA (siRNA) resulted in 50% decreased IER3 promoter activity and its mRNA level of the controls (Figure 1B and Supplementary Figure S1C). Endogenous expression of TAp73α was not readily detected in HeLa cells by western blot analysis (Figure 1B), implying that TAp73β but not TAp73α may play a significant role in cervical carcinoma cells. In order to identify the TAp73β-binding element in the IER3 promoter, we constructed serially truncated IER3 reporter plasmids as shown in Figure 1C. Luciferase reporter assay results showed that TAp73β retained transcriptional activity with truncated forms (-754, -561 and -283 bp) of IER3, but no transactivation was observed using the shorter construct (-69 bp) or the construct in which the p53-binding element was deleted (Δ-246–-218) (Figure 1C), indicating that TAp73β binds to the p53 consensus motif upstream of the IER3 promoter.
IER3 is a novel target gene of TAp73β.
(A) Promoter activation of IER3 by p53 family proteins was determined by luciferase reporter assay after transfection with increasing amounts of plasmids (50, 100, or 200 ng) encoding HA-tagged TAp73β, Flag-tagged TAp63 and HA-tagged p53 in HeLa, KB, Caski, SiHa and 293T cells. The luciferase activity was analyzed 24 h after transfection. Overexpression of TAp73β, TAp63, p53 was confirmed by immunoblot analysis using anti-HA or anti-Flag antibodies. The full-blots membrane was cut into pieces according to estimated molecular weight of proteins of interest and probed with indicated antibodies. All cropped bolts have been run under the same experimental condition. (B) Reduced transcriptional activity of IER3 in TAp73β knockdown (100 and 200 nM) HeLa cells is presented. Efficient silencing of TAp73β using its specific siRNA (100 or 200 nM) was confirmed by western blot analysis. GAPDH was used as a loading control. (C) The indicated truncated and deletion mutants of the IER3 promoter constructs were cotransfected with HA-TAp73β expression plasmid (200 ng) in HeLa cells. Cells were harvested for luciferase assays 24 h after transfection. All of results are expressed as the mean ± SEM of three independent experiments performed in triplicate. Different letters denote statistically significant values (p < 0.05).
TAp73β binds to the p53 consensus motif of the IER3 promoter
To confirm the sequences of IER3 required for TAp73β binding, nuclear extracts of HeLa cells transfected with HA-tagged TAp73β or p53 were prepared for EMSA. As shown in Figure 2A, incubation of the TAp73β-overexpressing nuclear fraction with radiolabeled oligonucleotides corresponding to the p53 consensus element (-246–-218) yielded a clear complex formation that disappeared upon the addition of extra cold probe (lanes 6 vs 7). The complex formation was also detectable without the TAp73β overexpression (lanes 2 and 4), indicating that this DNA binding association is physiologically relevant. In contrast, binding of p53 to the IER3 promoter sequences was not observed, in agreement with the luciferase reporter assays (Figure 2A). These differential binding patterns were further confirmed in vivo by ChIP assay. TAp73β was strongly recruited of IER3 promoter, whereas p53 did not show any significant enrichment of IER3 DNA in HeLa cells (Figure 2B). On the contrary, 293T cells showed the opposite results, with p53 protein strongly recruited to the IER3 promoter compared to TAp73β (Figure 2B), which supports the differential IER3 promoter activations by TAp73β and p53 observed in Figure 1A. Furthermore, the increased expression of IER3 protein and its mRNA induced by TAp73β but not by p53 was also confirmed (Figure 2C and Supplementary Figure S1D) and depletion of TAp73β indeed decreased the level of endogenous IER3 protein in HeLa cells (Figure 2D). Thus, these results indicate that TAp73β specifically binds to the p53 binding element of the IER3 promoter and modulates its transcription, but this does not happen with p53 in cervical cancer cells.
TAp73β binds to the IER3 promoter and regulates IER3 expression.
(A) The sequences of the IER3 probe encompassing the p53-binding element used to generate radiolabeled double-stranded oligonucleotides are shown. EMSA was performed using nuclear extracts (5 µg) isolated from HeLa cells overexpressing HA-TAp73β or -p53. No nuclear extract was incubated in lane 1. For cold probe, a 200 times excess of unlabeled oligonucleotides were used. Overexpression of HA-TAp73β or -p53 proteins and efficient nuclear subcellular fractionation were determined by immunoblotting using the indicated antibodies. (B) Both HeLa and 293T cells were transfected with plasmids encoding HA-TAp73β or -p53. The ChIP assay was performed using IER3-specific primers that targeted the p53-binding element and quantitative real-time RT-PCR results are shown as enrichment in fold. As a negative control, control IgG was used for immunoprecipitation. Asterisks indicate significant values compared with the control and the results are from three independent experiments run in duplicate (p < 0.05). HeLa cells were transfected with plasmids encoding HA-TAp73β or -p53 (C) and sip73 (D) for 24 h and cell lysates were prepared and immunoblotted with indicated antibodies. Quantitative analyses of the IER3 protein levels induced by TAp73β were shown as the means ± SEM of three independent experiments (bottom panel). GAPDH was used as an equal loading control. (A, C and D) These full-blots membrane was cut into pieces according to estimated molecular weight of proteins of interest and probed with indicated antibodies. All cropped bolts have been run under the same experimental condition.
IER3 mediates TAp73β-induced cell death
In order to determine the functional role of IER3 in cervical cancer cells, we assessed cell viability. Ectopic expression of IER3 promoted cell death (Figure 3A), while its knockdown enhanced survival of HeLa cells (Figure 3B). A similar trend was also observed in other HPV-positive cell lines including KB, Caski and SiHa cells (Supplementary Figures S2A and S2B). Ectopic expression of TAp73β or p53 efficiently induced death of HeLa cells to a similar extent (Figure 3C). However, TAp73β-induced cell death was significantly compromised in IER3-silenced cells, while IER3 knockdown did not affect p53-mediated cell death (Figure 3C), suggesting that IER3 could be a specific mediator of cell death induced by TAp73β, but not p53. In addition, the population of TAp73β-induced Annexin V-positive apoptotic cells (Figure 3D) and TAp73β-induced activation of Caspase 9, 7 and 3 (Figure 3E) were significantly reduced in IER3-depleted HeLa cells. In contrast, activation of Caspase 8 is not likely involved in this apoptotic signaling (Figure 3E).
IER3 is a mediator of TAp73β-induced apoptosis.
HeLa cells were transfected with plasmids encoding FLAG-IER3 (10, 30 and 50 ng) (A), siRNA for IER3 (50, 100 and 200 nM) (B) and TAp73β- or p53-expression construct in the presence or absence of siIER3 (C) and cell viability was measured at 24 h after transfection. (D) The proportion of Annexin V–positive apoptotic cells was analyzed by flow cytometry after cotransfection with TAp73β and scrambled or IER3-specific siRNA. Results are expressed as the means ± SEM of three independent experiments performed in triplicate. The statistically significant values are indicated with different letters or asterisks (p < 0.05). (E) HeLa cells were cotransfected with TAp73β and scrambled or IER3-specific siRNA and harvested 24 h after transfection. Activation of Caspase 3, 7, 8 and 9 was determined by western blot analysis using respective antibodies. (A, B and E) These full-blots membrane was cut into pieces according to estimated molecular weight of proteins of interest and probed with indicated antibodies. All cropped bolts have been run under the same experimental condition.
TAp73β-mediated IER3 upregulation is necessary for etoposide-induced cell death
Moreover, we treated HeLa cells using agents that may promote DNA damage-induced apoptosis and assessed the change in TAp73β and p53 levels. In general, these agents, including UV, nocodazole, camptothecin, doxorubicin, etoposide and cisplatin, upregulated TAp73β or p53. However, only limited agents simultaneously upregulated both TAp73β and IER3 in HeLa cells (Supplementary Figure S3A). In particular, well-known DNA damaging agents etoposide (200 μM) and doxorubicin (2 μM) significantly induced TAp73β protein expression by 7–9-fold, but only etoposide concomitantly increased the level of IER3 (Figure 4A), implying a functional role of IER3 in mediating etoposide signaling. Indeed, the IER3 knockdown cells were significantly resistant to etoposide-induced cell death, while they remained sensitive to doxorubicin-induced killing (Figure 4B). Furthermore, concentration-dependent cell death in response to etoposide was greatly inhibited by silencing of either TAp73β or IER3, whereas the silencing did not affect doxorubicin-induced cell death at all (Figures 4C and D). Etoposide-induced Annexin V-positive apoptotic cells were also decreased by silencing of either p73β or IER3 (Supplementary Figure S3B), while the knockdown did not affect doxorubicin-induced apoptosis (Supplementary Figure S3C). IER3 induction after etoposide treatment is suggested to be mediated by TAp73β because etoposide failed to increase IER3 expression in TAp73β-depleted cells (Figure 4E). In addition, IER3 is likely a key downstream mediator that is required for etoposide-induced cell death, as the partially inhibited etoposide activity after TAp73β depletion was fully recovered upon the knock-in of IER3 in the TAp73β-silenced cells (Figure 4F). Consistent results were observed by flow cytometary analysis of Annexin V-positive apoptotic cells (Supplementary Figure S3D).
Etoposide-induced apoptosis of cervical cancer cells is mediated by TAp73β and IER3.
(A) HeLa cells were incubated with 0.1% dimethylsulfoxide (DMSO), etoposide (ETO, 200 μM), or doxorubicin (DOXO, 2 μM) for 24 h. Representative immunoblot (left panel) and quantitative analysis of the TAp73β and IER3 levels are shown (right panel). Symbols (* and #) indicate significant values compared with the respective solvent controls and the results represent three independent experiments run in triplicate (p < 0.05). (B) The HeLa cells were transfected with scrambled or IER3-specific siRNAs, cells were treated with DMSO, ETO, or DOXO for 24 h and their viability was measured. The HeLa cells were transfected with scrambled, p73-specific (C), or IER3-specific (D) siRNAs, they were incubated with increasing concentrations of ETO (left panel) or DOXO (right panel) for 24 h and cell viability was measured. (E) HeLa cells were transfected with scrambled or p73-specific siRNA and then exposed to DMSO, ETO, or DOXO for 24 h. Using cell lysates, changes in the expression level of IER3 were determined by immunoblot analysis. Quantitative analysis of the IER3 levels is shown in the right panel (n = 3). (F) HeLa cells were transfected with the IER3-expressing plasmid and p73-specific siRNA as indicated and treated with DMSO or ETO and their cellular viability was measured. All results are expressed as the mean ± SEM of three independent experiments performed in triplicate. Statistically significant values are indicated with different letters or asterisks (p < 0.05). (A and E) These full-blots membrane was cut into pieces according to estimated molecular weight of proteins of interest and probed with indicated antibodies. All cropped bolts have been run under the same experimental condition.
c-Abl-mediated stimulation of TAp73β is critical for etoposide-mediated cell death
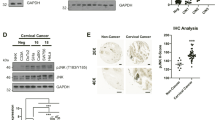
In order to elucidate how etoposide upregulates TAp73β in cervical carcinoma cells, we assessed the change in TAp73β mRNA level and found that its transcript level was not altered by etoposide treatment (Supplementary Figure S4A). Thus, next, we investigated whether etoposide-induced upregulation of TAp73β involved tyrosine kinase c-Abl, which is known to stimulate p73 activity by increasing stability26. Using a specific inhibitor of c-Abl, imatinib (STI571; Gleevec), alteration of etoposide-mediated TAp73β protein level was determined. As shown in the Figure 5A, etoposide increased the phosphorylation of Tyr 99 residue of etoposide failed to increase the level of TAp73β and IER3 in the presence of imatinib, while imatinib had no effect on doxorubicin-mediated regulation of TAp73β. This failure of etoposide to upregulate TAp73β and IER3 was also confirmed by knockdown of c-Abl using a siRNA specific to c-Abl (Supplementary Figure S4B). In addition, TAp73β-mediated transcriptional activation of IER3 was diminished upon c-Abl inhibition (Figure 5B). Similarly, etoposide-induced cell death was significantly inhibited by imatinib (Figure 5C) and c-Abl knockdown (Figure 5D) and the inhibitory action of imatinib on etoposide-mediated cell death induced was overcome by ectopic expression of either TAp73β or IER3 (Figure 5E and Supplementary Figure S4C). Thus, together these results suggest that the activation of TAp73β by c-Abl tyrosine kinase leads to the upregulation of IER3 and is an important signaling axis in etoposide-induced apoptosis of HeLa cells.
Etoposide-induced upregulation of TAp73β and IER3 is mediated by c-Abl.
(A) HeLa cells were incubated with DMSO, ETO, or DOXO for 24 h in the presence or absence of imatinib (10 μM). The changes in the level of TAp73β and IER3 were shown by immunoblotting (left panel) and quantified results are presented in the right panel. The full-blots membrane was cut into pieces according to estimated molecular weight of proteins of interest and probed with indicated antibodies. All cropped bolts have been run under the same experimental condition. (B) HeLa cells were cotransfected with TAp73β-encoding plasmid and IER3 promoter construct and incubated with or without imatinib for 24 h. Then, luciferase activity was measured using the cell lysates. (C) HeLa cells were incubated with increasing concentrations of ETO in the presence or absence of imatinib for 24 h and cell viability was measured. (D) HeLa cells were transfected with scrambled or c-Abl siRNAs (200 nM). Following transfection for 12 h, the cells were incubated with ETO for 24 h and cell viability was measured. (E) HeLa cells were transfected with IER3- and/or TAp73β-expression plasmids with or without ETO and/or imatinib and cell viability was measured after 24 h incubation. All results are expressed as the means ± SEM of three independent experiments performed in triplicate. Statistically significant values are indicated with different letters or asterisks (p < 0.05).
Expression of TAp73β and IER3 is downregulated in cervical cancer patients
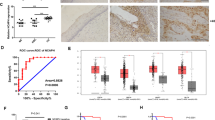
Since our molecular and cellular experimental results suggested that IER3 induction by upregulated TAp73β is crucial for apoptosis of cervical cancer cells, we examined the expression profiles of TAp73β and IER3 in HPV-infected cervical cancer patients. Western blot analysis of the epithelium isolated from cervical tissues of women free of cancer showed clear expression of both TAp73β and IER3 proteins with correlation coefficient of 0.83 (Figure 6A). In sharp contrast, the expression levels of p73β and IER3 proteins were extremely low or undetectable in cervical cancer tissues (Figure 6A). The expression of their mRNAs was also significantly downregulated in cervical cancer tissues (Figure 6B). In accordance, immunohistochemical analysis of normal and cervical carcinoma tissues supported the western blot results, in which both proteins showed positive nuclear staining in normal cervical epithelium but not in cancer tissues (Figure 6C).
Lack of the expression of TAp73β and IER3 was observed in cervical carcinoma tissues.
(A) Equal amounts of extracted protein from FFPE tissues of normal human cervical epithelium (n = 9) and cervical carcinoma tissues (n = 10) were subjected to SDS-PAGE for immunoblot analysis using indicated antibodies (upper panel). The full-blots membrane was cut into pieces according to estimated molecular weight of proteins of interest and probed with indicated antibodies. All cropped bolts have been run under the same experimental condition. Areas of cervical tissues scrapped for protein extraction are indicated with dotted lines (lower panel). The relative quantified expression of both p73β and IER3 proteins between the control cervix and cervical cancer were compared. Estimated regression line superimposed on scatter plot of levels of p73β and IER3 proteins in normal cervix is also presented with a correlation coefficient. (B) The total mRNAs from FFPE tissues of control human cervix (n = 5) and cervical carcinoma (n = 5) were extracted and used for a quantitative real-time PCR analysis of p73 and IER3. (C) Representative immunohistochemistical analyses of p73 and IER3 expression in control cervix and cervical cancer are shown.
Discussion
The accumulation of p53 after DNA-damage has been considered an important step in inducing apoptosis and cell cycle arrest of various cancers upon chemotherapy and radiotherapy27. However, p53-based gene therapy to restore its expression often fails to inhibit cervical cancer10 because E6 oncoprotein, produced by high-risk HPVs, recruits E6-associated protein (E6AP), an E3 ligase and forms a trimeric complex with p53, resulting in ubiquitination and proteosomal degradation of p537,9,28. In contrast, because p73 and E6 protein do not interact8, p73 is an important and promising molecule that may inhibit the growth of cervical cancer9,10,11,29,30,31,32.
Here, we identified IER3, for the first time, as a critical and specific mediator of TAp73β function in HPV-positive human cervical cancer. TAp73β associates with the IER3 promoter at the known p53 consensus sequence and stimulates IER3 transcription in cervical carcinoma cells (Figures 1 and 2). However, p53 exhibited no transcriptional activity on IER3 in HPV-infected cervical cancer cells, including HeLa, KB, Caski and SiHa cells (Figures 1 and 2) even though the present and previous studies showed that p53 can transcriptionally regulate human IER3 in other types of cells, such as the observed activation in human embryonic kidney (293T) and hepatoma (Hep3B) cell lines14 and repression in human colorectal carcinoma (HCT116 and SW480) and fibroblast cell lines (HaCaT)16 (Supplementary Figure S1B). The distinctive lack of p53 activity in cervical cancer cells reflects its failure to associate with the IER3 promoter, as evident by EMSA and ChIP (Figures 2A and B). Understanding the mechanism underscoring this discriminative effect by TAp73β and p53 in different cellular contexts requires further studies. However, other factors that associate with p53 in HPV-infected cervical cancer cells may possibly hinder its binding to the p53-responsive element in IER3 sequences.
Differential transcriptional activities of the p73 protein on its target genes such as BAX, NOXA and NIS have been observed in different cell types by others33,34,35. In the present study, we have also found that TAp73β distinctively regulates IER3 in cervical cancer cells. This discriminatory effect of TAp73β on IER3 regulation in different cellular contexts that we have found in this study is likely a consequence of TAp73β's differential ability to associate with the IER3 promoter, as TAp73β failed to associate with the IER3 promoter and was unable to stimulate transcription of IER3 in 293T cells (Figures 1A and 2B). Although a detailed understanding of the underlying regulatory molecular mechanism responsible for the differential effects exerted by TAp73β has yet to be achieved, we can speculate the potential involvement of distinct cellular factors, such as transcriptional co-activators of TAp73 that allow TAp73 access to its binding element in the IER3 promoter.
At present, little information is available about how TAp73β inhibits the growth of cervical cancer cells. In this study, we discovered that functional IER3 is necessary and integral to TAp73β-mediated apoptotic activity in cervical cancer cells (Figure 3). In addition, we obtained a striking result, that the expression levels of both TAp73β and IER3 proteins were undetectable in cervical carcinomas, whereas the two proteins were more abundantly expressed in the epithelium of normal human cervix (Figure 6A). To our knowledge, this is the first report that showed this striking difference in the level of TAp73β and IER3 proteins between cervical carcinoma and normal cervical epithelium by quantitative western blot analysis. Additionally, TAp73β and IER3 expression correlated, supporting the idea that IER3 is probably a pathophysiologically relevant downstream target that acts as a mediator of TAp73β-induced apoptosis in human cervix. In addition, the undetectable expression levels of TAp73β and IER3 proteins in cervical cancer patients implies that the two proteins likely act as tumor suppressors by preventing aberrant proliferation and can also serve as molecular markers to discriminate cervical carcinoma from other cancers. At present, the role of IER3 in tumorigenesis is not clearly understood and only limited studies have been performed that examine the expression levels of IER3 in different types of cancer. The proposed role of IER3 is even contradictory. It is increased in multiple myeloma and colorectal carcinoma, while decreased in ovarian carcinoma and positive IER3 expression is associated with better prognosis of pancreatic adenocarcinoma patients36,37,38,39,40. This variable profile of IER3 expression in cancer is likely reflective of contrasting functions of IER3 in different cellular contexts, acting as a pro-apoptotic or an anti-apoptotic molecule.
DNA damage-induced upregulation of p73 and subsequent apoptosis of cancer cells in response to radiotherapy and chemotherapeutics, including etoposide, doxorubicin, cisplatin, camptothecin, nocodazole and taxol has been reported11,30,41,42,43,44. In this study, we, for the first time, identified that TAp73β-induced IER3 expression is a crucial mediator of etoposide-induced death in cervical cancer cells (Figure 4). Etoposide is a widely used chemotherapeutic for many cancers, including cervical carcinoma, because of its ability to break DNA strands by forming a ternary complex with topoisomerase 2 and DNA, leading to the apoptosis of cancer cells45,46,47,48,49. The TAp73β-induced IER3 expression specifically correlated with chemosensitivity to etoposide (Figure 4), suggesting that the ability of cervical cancer cells to upregulate TAp73β and IER3 proteins is a critical factor for responsiveness to etoposide. In addition, etoposide-induced apoptotic cell death was regulated by c-Abl tyrosine kinase signaling, as etoposide-mediated apoptosis and upregulation of TAp73β protein was effectively inhibited by the c-Abl inhibitor imatinib or the c-Abl knockdown (Figure 5 and Supplementary Figure S5B). c-Abl is an important kinase that mediates apoptotic cell death and cell cycle arrest50. When DNA damage has occurred, c-Abl is activated by its phosphorylation and phosphorylates p73 isoforms at Tyr 9926, resulting in the stabilization of p73 proteins51. Therefore, c-Abl-mediated upregulation of TAp73β and subsequent transactivation of IER3 confers chemosensitivity of cervical cancer cells to etoposide. Of interest, our findings also suggest that the use of imatinib, which is also used to treat multiple cancers, should be avoided for cervical cancer patients who undergo etoposide chemotherapy, as it prevents etoposide-induced upregulation of TAp73β and IER3.
In summary, we identified that, in cervical carcinoma cells, IER3 was distinctively regulated by TAp73β, unlike in other types of cells where p53 is the main regulator and the c-Abl/TAp73/IER3 signaling axis controlled etoposide-induced death of cervical cancer cells.
Methods
Cells culture and reagents
HeLa, SiHa and 293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. RPMI1640 was used to culture CaSki, SK-OV-3, HCT116 and SW480 cells. KB cells were cultured in RPMI1640 media that contain 25 mM HEPES and sodium bicarbonate. Cells were grown in an incubator at 37°C with 5% CO2. Reagents used for cell culture were purchased from Caisson (Caisson, North Logan, UT, USA). The anti-p73 (558787, BD Biosciences, San Jose, CA, USA), anti-HA (H6908, Sigma-Aldrich, St. Louis, MO, USA) and anti-α-tubulin (LF-PA0146, AbFrontier Seoul, Korea) were purchased for experiments. The anti-p53 (sc-126), anti-IER3 (sc-33171), anti-PARP (sc-74469) and anti-GAPDH (sc-25778) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The anti-Caspase 3 (9662), 8 (9746), 9 (9508), anti-FLAG (2368) and anti-p-p73 (4665) antibodies were purchased from Cell Signaling (Danvers, MA, USA). Caspase 7 antibody was purchased from NOVUS Biologicals (NB100-56529, Littleton, CO, USA). Etoposide, camptothecin and cisplatin were purchased from Calbiochem (San Diego, CA, USA) and imatinib was obtained from Santa Cruz Biotechnology. Doxorubicin, nocodazole, sodium vanadate (Na3VO4) and sodium fluoride (NaF) were purchased from Sigma-Aldrich. Padexol (taxol) was obtained from Shin Poong Pharm. Co (Seoul, Korea). The protease inhibitor cocktail was purchased from GenDEPOT (Barker, TX, USA).
Plasmid constructs
The human IER3 promoter (-1384 – +74) was generated by recombinant PCR using extracted blood genomic DNA as a template and the following primers: p(-1384)-F (5′-GGGACGCGTCTCCTGAGCTCAAGT) with p(+75)-R (5′-GATGGTCATGGTCGGGTGGCA) and p(+75)-F (5′-TGCCACCCGACCATGACCATCATGGAAGACGCCAAAAACATA) with SphI-R (5′-ATCTCTGGCATGCGAGAATCT). The truncated p(-754), p(-283), p(-69) promoters of IER3 were PCR amplified using following primers: -754-F (5′-GGGACGCGTGGGTCAGTATTGCAGCAGGAT) with SphI-R, -283-F (5′- GGGACGCGTCCTGTGAGGGATCCTGTGGCT) with SphI-R and -69-F (5′- GGGACGCGTTGCGGGAGGAGGAGTTAGAAG) with SphI-R. The deletion construct of the IER3 promoter was generated by recombinant PCR using following primers: p(-1384)-F with p(-1384mut)-R (5′-CCGGGCTGCAGACCTGAG) and p(-1384mut)-F (5′- CCTCTCCAGGTCTGCAGC). The PCR products were digested with MluI and SphI (Enzynomics, Seoul, Korea) and ligated into the pGL3 basic vector (Clontech, Mountain View, CA, USA). The FLAG-tagged IER3 expression plasmid was produced by PCR amplification using the following primers: IER3-F (5′-GCCTCCGGATCCATGGACTACAAAGACGACGACGACAAATGTCACTCTCGCAGC) and IER3-R (5′-GAAGCCGAATTCTTAGAAGGCGGCCGGGT). The PCR products were digested with BamHI and XhoI (Enzynomics) and ligated into pcDNA3 (Invitrogen, Carlsbad, CA, USA). The pcDNA3 HA-tagged TAp73β and p53 plasmids were generous gifts from Dr. Kyung Hee Choi (Chung-Ang University, Seoul, Korea). Human TAp63 cDNA was purchased from Thermo Scientific (Rockford, IL, USA) and amplified using following primers: TAp63-F (5′-GACGGATCCATGGACTACAAAGACGACGACGACAAAAATTTTGAAACTTCACG) with TAp63-R (5′-GCTCGAGCGGCCGCTCACTCCCCCTCCTCTTTGA). The product was digested with BamHI and NotI (Enzynomics) and cloned into pcDNA3.
Luciferase assay
293T, KB and SiHa cells (5 × 104) and other cell lines (2 × 104) were transfected with 100 ng of IER3-luciferase reporter, 50 ng of pCMV ß-galactosidase (Clontech) and indicated amounts of plasmids encoding TAp73β, TAp63, or p53 using Neon transfection system (Invitrogen) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were then incubated in 12 or 48-well plates containing medium for 24 h. Luciferase activity was assessed as previously described52. Absorbances were measured with the FlexStation3 Microplate Reader (Molecular Devices, Sunnyvale, CA, USA).
Subcellular fractionation
HeLa cells (1 × 106) were transfected with expression plasmids and fractionation of nuclear and cytosolic compartments was performed as reported previously53.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as previously described52. Double-stranded oligonucleotides of human IER3 sense (5′-AGGTGCCACATGCCTCGACATGTGCCTG) and antisense (5′-CAGGCACATGTCGAGGCATGTGGCACCT) were annealed before use.
Chromatin immunoprecipitation (ChIP) analysis
ChIP assays were performed as previously described52. DNA was amplified using the following primer set flanking the TAp73β binding element in the IER3 promoter: forward (5′-CCTGTGAGGGATCCTGTGGC) and reverse (5′-AGTGGGTGGAGACTTGACAT). Products were analyzed by quantitative real-time PCR.
Cell viability assay
HeLa cells (2 × 104) were transfected using the Neon system (Invitrogen). Cell viability was measured by CellTiter-Glo Assay (Promega, Madison, WI, USA), according to the manufacturer's instructions.
Flow cytometry analysis
Annexin V-positive apoptotic cells were detected as previously reported54.
Immunoblot analysis
HeLa cells (1 × 106) were transfected with the indicated plasmids as well as small interference nucleotides (siRNAs). After a 24-h transfection, cell lysates were prepared and subjected to SDS-PAGE for immunoblotting with respective antibodies. The membranes were detected using a ChemiDoc™ XRS+ System Imager (Bio-Rad Laboratories, Hercules, CA, USA) and the intensity of each band was quantified using Quantity One software (Bio-Rad Laboratories).
RNA interference
Small-interfering RNA (siRNA) target sequences against p73 and IER3 were 5′- CGGAUUCCAGCAUGGACGU and 5′-CCAGCCAAAAGGCUUCUCUUU, respectively. The control siRNA sequence used was 5′-CCUACGCCACCAAUUUCGU. The sense and antisense oligonucleotides were annealed in the presence of Annealing Buffer (Bioneer, Daejeon, South Korea). A siRNA specific to c-Abl was purchased from Santa Cruz Biotechnology.
Human subjects and cervical tissues
Formalin-fixed paraffin-embedded (FFPE) block sections (15 μm thick) of cervical tumors from 10 patients (mean age = 47.7) and control cervical tissues from 9 women (mean age = 49.2) diagnosed with uterine myoma were examined. The sections were reviewed by pathologists and obtained from the Bio-Resource Center at the Seoul Asan Medical Center. The present study was reviewed and approved by the Seoul Asan Medical Center Institutional Review Boards. Informed consent was obtained from all subjects participated in this study. The methods were carried out in accordance with the approved guidelines.
Protein extraction from human cervical tissues
Total proteins were extracted from the epithelial layer of normal cervical tissue and cervical cancer sections of FFPE tissue sections. The FFPE tissues were deparaffinized with xylene (Duksan, Ansan, Korea) at room temperature for 10 min three total times. Then, tissue protein extracts were retrieved and pelleted by centrifugation at 14,000 × g for 2 min and the supernatant was carefully removed. The deparaffinized tissue pellets were then rehydrated with a graded series of ethanol (EMD Millipore Corp, Billerica, MA, USA), 100% for 5 min and then two repeated minutes with 95%, 90%, 80% and 70%. The rehydrated tissue sections were resuspended in extraction buffer (RIPA buffer with 2% SDS, 1 mM Na3VO4, 10 mM NaF and protease inhibitor cocktail). The samples were mixed by vortexing and boiled at 100°C for 20 min, followed by the incubation at 80°C in a heat block for 2 h. During the incubation, samples were briefly vortexed every 20 min. At the end of the incubation, protein extracts were collected by centrifugation (14,000 × g) for 30 min at 4°C and then quantitated using a BCA protein assay kit (Pierce Chemicals Co, Rockford, IL, USA).
RNA extraction and real-time PCR analysis
Total RNAs from cervical tissues and other cell lines were isolated by the PureLinkTM FFPE total RNA isolation kit (Invitrogen) and TRIzol reagent (Invitrogen), following the manufacturer's instructions. The concentration and quality of RNA were determined with an ND-1000 spectrophotometer (NanoDrop, Waltham, MA, USA). Reverse-transcription to cDNA was performed using the SuperScriptIII first-strand synthesis kit (Invitrogen). All cDNAs used in real-time PCR were normalized with GAPDH. Quantitative real-time PCRs were performed using an iQTM SYBR Green Supermix (Bio-Rad Laboratories). Gene expression was quantified by the delta-delta-CT method and real-time PCRs were performed in a CFX-96TM thermal cycler and detection system (Bio-Rad Laboratories). The nucleotide sequences of primers used for real-time PCR (Bioneer) are: p73-F (5′-GACGAGGACACGTACTACCTT) and p73-R (5′-CTGCCGATAGGAGTCGACCA); IER3-F (5′-CAGCCGCAGGGTTCTCTAC) and IER3-R (5′-GATCTGGCAGAAGACGATGGT); GAPDH-F (5′-AGGGGCCATCCACAGTCTT) and GAPDH-R (5′- AGCCAAAAGGGTCATCATCTCT).
Haematoxylin and eosin staining and immunohistochemistry
Immunohistochemistry were performed according to our previous study55. The sections were incubated with anti-human p73 (1:50) or anti-human IER3 (1:50) in antibody diluent (Dako, Carpinteria, CA, USA) for 24 h at 4°C.
Statistical analysis
Multiple comparison analyses of values were performed with the Student-Newman-Keuls test using SAS version 9.2 (SAS Institute, Cary, NC, USA) and Student's t-test was used for comparisons with control. The data are presented as means ± SEM and p < 0.05 was considered to be statistically significant.
References
Ferlay, J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 127, 2893–2917 (2010).
Munoz, N., Castellsague, X., de Gonzalez, A. B. & Gissmann, L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 24 Suppl 3S3/1–10 (2006).
Bernard, H. U. et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 401, 70–79 (2010).
Moody, C. A. & Laimins, L. A. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 10, 550–560 (2010).
Collavin, L., Lunardi, A. & Del Sal, G. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 17, 901–911 (2010).
Werness, B. A., Levine, A. J. & Howley, P. M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 248, 76–79 (1990).
Scheffner, M., Werness, B. A., Huibregtse, J. M., Levine, A. J. & Howley, P. M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 63, 1129–1136 (1990).
Marin, M. C. et al. Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol. 18, 6316–6324 (1998).
Das, S., El-Deiry, W. S. & Somasundaram, K. Efficient growth inhibition of HPV 16 E6-expressing cells by an adenovirus-expressing p53 homologue p73beta. Oncogene. 22, 8394–8402 (2003).
Das, S. & Somasundaram, K. Therapeutic potential of an adenovirus expressing p73 beta, a p53 homologue, against human papilloma virus positive cervical cancer in vitro and in vivo. Cancer Biol Ther. 5, 210–217 (2006).
Liu, S. S. et al. Enhancement of the radiosensitivity of cervical cancer cells by overexpressing p73alpha. Mol Cancer Ther. 5, 1209–1215 (2006).
Arlt, A. & Schafer, H. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol. 90, 545–552 (2011).
Kumar, R. et al. A novel immediate early response gene, IEX-1, is induced by ultraviolet radiation in human keratinocytes. Biochem Biophys Res Commun. 253, 336–341 (1998).
Schafer, H., Diebel, J., Arlt, A., Trauzold, A. & Schmidt, W. E. The promoter of human p22/PACAP response gene 1 (PRG1) contains functional binding sites for the p53 tumor suppressor and for NFkappaB. FEBS Lett. 436, 139–143 (1998).
Huang, Y. H., Wu, J. Y., Zhang, Y. & Wu, M. X. Synergistic and opposing regulation of the stress-responsive gene IEX-1 by p53, c-Myc and multiple NF-kappaB/rel complexes. Oncogene. 21, 6819–6828 (2002).
Im, H. J., Pittelkow, M. R. & Kumar, R. Divergent regulation of the growth-promoting gene IEX-1 by the p53 tumor suppressor and Sp1. J Biol Chem. 277, 14612–14621 (2002).
Yoon, S. et al. IEX-1-induced cell death requires BIM and is modulated by MCL-1. Biochem Biophys Res Commun. 382, 400–404 (2009).
Sebens Muerkoster, S. et al. The apoptosis-inducing effect of gastrin on colorectal cancer cells relates to an increased IEX-1 expression mediating NF-kappa B inhibition. Oncogene. 27, 1122–1134 (2008).
Yamashita, K., Nakashima, S., You, F., Hayashi, S. & Iwama, T. Overexpression of immediate early gene X-1 (IEX-1) enhances gamma-radiation-induced apoptosis of human glioma cell line, U87-MG. Neuropathology. 29, 20–24 (2009).
Arlt, A. et al. The early response gene IEX-1 attenuates NF-kappaB activation in 293 cells, a possible counter-regulatory process leading to enhanced cell death. Oncogene. 22, 3343–3351 (2003).
Ustyugova, I. V., Zhi, L., Abramowitz, J., Birnbaumer, L. & Wu, M. X. IEX-1 deficiency protects against colonic cancer. Mol Cancer Res. 10, 760–767 (2012).
Wu, M. X., Ao, Z., Prasad, K. V., Wu, R. & Schlossman, S. F. IEX-1L, an apoptosis inhibitor involved in NF-kappaB-mediated cell survival. Science. 281, 998–1001 (1998).
You, F., Osawa, Y., Hayashi, S. & Nakashima, S. Immediate early gene IEX-1 induces astrocytic differentiation of U87-MG human glioma cells. J Cell Biochem. 100, 256–265 (2007).
Pawlikowska, P. et al. ATM-dependent expression of IEX-1 controls nuclear accumulation of Mcl-1 and the DNA damage response. Cell Death Differ. 17, 1739–1750 (2010).
Arlt, A. et al. IEX-1 directly interferes with RelA/p65 dependent transactivation and regulation of apoptosis. Biochim Biophys Acta. 1783, 941–952 (2008).
Yuan, Z. M. et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 399, 814–817 (1999).
Lu, C. & El-Deiry, W. S. Targeting p53 for enhanced radio- and chemo-sensitivity. Apoptosis. 14, 597–606 (2009).
Scheffner, M., Huibregtse, J. M., Vierstra, R. D. & Howley, P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 75, 495–505 (1993).
Nenutil, R., Ceskova, P., Coates, P. J., Nylander, K. & Vojtesek, B. Differential expression of p73alpha in normal ectocervical epithelium, cervical intraepithelial neoplasia and invasive squamous cell carcinoma. Int J Gynecol Pathol. 22, 386–392 (2003).
Liu, S. S. et al. p73 expression is associated with the cellular radiosensitivity in cervical cancer after radiotherapy. Clin Cancer Res. 10, 3309–3316 (2004).
Wakatsuki, M. et al. p73 protein expression correlates with radiation-induced apoptosis in the lack of p53 response to radiation therapy for cervical cancer. Int J Radiat Oncol Biol Phys. 70, 1189–1194 (2008).
Lee, J. J., Kim, S., Yeom, Y. I. & Heo, D. S. Enhanced specificity of the p53 family proteins-based adenoviral gene therapy in uterine cervical cancer cells with E2F1-responsive promoters. Cancer Biol Ther. 5, 1502–1510 (2006).
Stros, M., Ozaki, T., Bacikova, A., Kageyama, H. & Nakagawara, A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J Biol Chem. 277, 7157–7164 (2002).
Grande, L. et al. Transcription factors Sp1 and p73 control the expression of the proapoptotic protein NOXA in the response of testicular embryonal carcinoma cells to cisplatin. J Biol Chem. 287, 26495–26505 (2012).
Guerrieri, F. et al. The sodium/iodide symporter NIS is a transcriptional target of the p53-family members in liver cancer cells. Cell Death Dis. 4, e807 (2013).
Ria, R. et al. Gene expression profiling of bone marrow endothelial cells in patients with multiple myeloma. Clin Cancer Res. 15, 5369–5378 (2009).
Segditsas, S. et al. Putative direct and indirect Wnt targets identified through consistent gene expression changes in APC-mutant intestinal adenomas from humans and mice. Hum Mol Genet. 17, 3864–3875 (2008).
Lee, S., Bang, S., Song, K. & Lee, I. Differential expression in normal-adenoma-carcinoma sequence suggests complex molecular carcinogenesis in colon. Oncol Rep. 16, 747–754 (2006).
Han, L. et al. Clinical significance of IEX-1 expression in ovarian carcinoma. Ultrastruct Pathol. 35, 260–266 (2011).
Sasada, T. et al. Prognostic significance of the immediate early response gene X-1 (IEX-1) expression in pancreatic cancer. Ann Surg Oncol. 15, 609–617 (2008).
Lin, K. W., Nam, S. Y., Toh, W. H., Dulloo, I. & Sabapathy, K. Multiple stress signals induce p73beta accumulation. Neoplasia. 6, 546–557 (2004).
Codelia, V. A., Cisterna, M., Alvarez, A. R. & Moreno, R. D. p73 participates in male germ cells apoptosis induced by etoposide. Mol Hum Reprod. 16, 734–742 (2010).
Tiwary, R., Yu, W., Sanders, B. G. & Kline, K. alpha-TEA cooperates with chemotherapeutic agents to induce apoptosis of p53 mutant, triple-negative human breast cancer cells via activating p73. Breast Cancer Res. 13, R1 (2011).
Al-Bahlani, S. et al. P73 regulates cisplatin-induced apoptosis in ovarian cancer cells via a calcium/calpain-dependent mechanism. Oncogene. 30, 4219–4230 (2011).
Bae, J. H. et al. Neoadjuvant cisplatin and etoposide followed by radical hysterectomy for stage 1B-2B cervical cancer. Gynecol Oncol. 111, 444–448 (2008).
Tanaka, T., Bai, T., Yukawa, K. & Umesaki, N. Optimal combination chemotherapy and chemoradiotherapy with etoposide for advanced cervical squamous cancer cells in vitro. Oncol Rep. 15, 939–947 (2006).
Meyer, T., Nelstrop, A. E., Mahmoudi, M. & Rustin, G. J. Weekly cisplatin and oral etoposide as treatment for relapsed epithelial ovarian cancer. Ann Oncol. 12, 1705–1709 (2001).
Lock, R. B., Thompson, B. S., Sullivan, D. M. & Stribinskiene, L. Potentiation of etoposide-induced apoptosis by staurosporine in human tumor cells is associated with events downstream of DNA-protein complex formation. Cancer Chemother Pharmacol. 39, 399–409 (1997).
El-Awady, R. A., Ali, M. M., Saleh, E. M. & Ghaleb, F. M. Apoptosis is the most efficient death-pathway in tumor cells after topoisomerase II inhibition. Saudi Med J. 29, 558–564 (2008).
Agami, R., Blandino, G., Oren, M. & Shaul, Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 399, 809–813 (1999).
Gong, J. G. et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 399, 806–809 (1999).
Park, M. et al. FOXL2 interacts with steroidogenic factor-1 (SF-1) and represses SF-1-induced CYP17 transcription in granulosa cells. Mol Endocrinol. 24, 1024–1036 (2010).
Kim, J. H. et al. FOXL2 posttranslational modifications mediated by GSK3beta determine the growth of granulosa cell tumours. Nat Commun. 5, 2936 (2014).
Kim, J. H. et al. Differential apoptotic activities of wild-type FOXL2 and the adult-type granulosa cell tumor-associated mutant FOXL2 (C134W). Oncogene. 30, 1653–1663 (2011).
Park, H. O. & Bae, J. Disturbed relaxin signaling pathway and testicular dysfunction in mouse offspring upon maternal exposure to simazine. PLoS One. 7, e44856 (2012).
Acknowledgements
The authors thank Ms. Seeun Park for the technical support with EMSA study. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Information Communication Technology (ICT) and Future Planning (2014R1A2A2A01006839) and by National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (1220090).
Author information
Authors and Affiliations
Contributions
H.J., D.S., T.K. and J.Y. prepared figures and analyzed the data. K.L. and J.B. supervised the project, analyzed the data and wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
IER3 is a crucial mediator of TAp73β-induced apoptosis in cervical cancer and confers etoposide sensitivity
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Jin, H., Suh, DS., Kim, TH. et al. IER3 is a crucial mediator of TAp73β-induced apoptosis in cervical cancer and confers etoposide sensitivity. Sci Rep 5, 8367 (2015). https://doi.org/10.1038/srep08367
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep08367
This article is cited by
-
Mild heat treatment in vitro potentiates human adipose stem cells: delayed aging and improved quality for long term culture
Biomaterials Research (2023)
-
TP73 Isoform-specific disruption reveals a critical role of TAp73beta in growth suppression and inflammatory response
Cell Death & Disease (2023)
-
TRAIL promotes hepatocellular carcinoma apoptosis and inhibits proliferation and migration via interacting with IER3
Cancer Cell International (2021)
-
Identification and validation of a six-gene signature associated with glycolysis to predict the prognosis of patients with cervical cancer
BMC Cancer (2020)
-
Mitochondria-targeting self-assembled nanoparticles derived from triphenylphosphonium-conjugated cyanostilbene enable site-specific imaging and anticancer drug delivery
Nano Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.