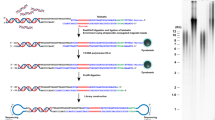
Abstract
Telomere length can provide valuable insight into telomeres and telomerase related diseases, including cancer. Here, we present a brand-new optical telomere length measurement protocol using surface enhanced Raman scattering (SERS). In this protocol, two single strand DNA are used as SERS probes. They are labeled with two different Raman molecules and can specifically hybridize with telomeres and centromere, respectively. First, genome DNA is extracted from cells. Then the telomere and centromere SERS probes are added into the genome DNA. After hybridization with genome DNA, excess SERS probes are removed by magnetic capturing nanoparticles. Finally, the genome DNA with SERS probes attached is dropped onto a SERS substrate and subjected to SERS measurement. Longer telomeres result in more attached telomere probes, thus a stronger SERS signal. Consequently, SERS signal can be used as an indicator of telomere length. Centromere is used as the inner control. By calibrating the SERS intensity of telomere probe with that of the centromere probe, SERS based telomere measurement is realized. This protocol does not require polymerase chain reaction (PCR) or electrophoresis procedures, which greatly simplifies the detection process. We anticipate that this easy-operation and cost-effective protocol is a fine alternative for the assessment of telomere length.
Similar content being viewed by others
Introduction
Telomeres, which contain tandem TTAGGG repeats, are a special DNA-protein complex located at the end of linear eukaryotic chromosomes1,2. Telomeres play a vital role in maintaining genome stability as they help to disguise and protect the chromosome ends3,4,5. Due to the end replication problem of linear chromosomes, telomeres are shortened each time when a cell divides6,7. After several cell division, telomeres are shortened to a critical length, which triggers the DNA damage response. Then the cell stops proliferating, gets into replicative senescence and eventually loses viability8,9,10. This is what happens in most human somatic cells. However, telomere length can be maintained in some other cell lines, for example, stem cells, germ cells and most importantly cancer cells. In 85% of the cancer cells, telomere length is sustained by a ribonucleoprotein complex named telomerase11. Telomerase can synthesize and elongate telomeric repeats using its own RNA component as the template12,13,14. While the remaining 15% of the cancer cells maintain their telomere length through alternative lengthening of telomere (ALT)15. Telomere maintenance provides these cells unlimited proliferative ability and immortality. Since telomeres of normal somatic cells are meant to undergo progressive shortening with increasing age, abnormal variation in telomere length is usually considered as a marked sign of the occurrence of diseases16. For example, some premature ageing syndromes show accelerated telomere loss while cancer cells exhibit no obvious telomere loss. As a consequence, measuring telomere length will provide important information for the diagnosis and therapeutics of human diseases17.
Conventional telomere length measurement method includes Southern blot18,19, quantitative PCR (qPCR)20,21, hybridization protection assay (HPA)22 and fluorescence in-situ hybridization (FISH)23,24. In southern blot, genome DNA is first digested by restriction endonuclease (e.g. Hinf I and Rsa I) to produce telomere/terminal restriction fragment (TRF). Then the digested genome DNA is separated by electrophoresis. TRF is visualized by hybridization with specific radioactive or fluorescent probes25. In HPA, genome DNA is directly hybridized with acridinium ester-labeled probe and telomere length is assessed by chemiluminescence signal22. In qPCR, PCR is introduced to amplify the target sequence, thus much less genome DNA is required in the assay26,27. While in FISH, fluorescent telomere specific peptide nucleic acid (PNA) probe is used to hybridize in-situ with telomere sequences in the cell nucleus28,29 and telomere length is measured using a fluorescence microscope. These methods have their own unique properties. For example, Southern blot is the most widely used method and is employed as the reference for HPA, FISH etc. It provides the absolute length of telomeres. HPA is fast and does not require highly purified and integrated genome DNA. qPCR only needs a small amount of cell/DNA sample. While telomere length acquired in other methods is an average of several hundreds (even millions) of cells, FISH can investigate telomeres in a single cell or a single chromosome29,30. Although these telomere length measurement strategies have made impressive progress, they still suffer from some unsatisfactory shortcomings, such as complicated operation procedures, relatively low accuracy and high cost. Consequently, it is still necessary to develop other telomere length measurement methods.
Considering all the mentioned above, here, a novel SERS based optical telomere length measurement protocol is presented. SERS is a fascinating phenomenon observed on nanoscale roughened metallic surfaces, where Raman scattering of adsorbed molecules is drastically boosted by the strong electromagnetic field around the metallic surfaces or charge transfer between molecules and the metal substrates31,32. SERS can enhance Raman signals by a factor of more than 10633,34. The excellent enhancing ability of SERS ensures an extremely high sensitivity of SERS based analysis, even on single molecule level35,36,37,38,39. The unique “fingerprint” spectral property of SERS makes it possible to track the analyte in complicated biological environments. As a result, SERS is widely used in biological detections40. The detection procedures of the presented SERS protocol (shown in Figure 1) can be described as follows: (1) Genome DNA is extracted from the cells. (2) Two SERS probes, i.e. telomere specific single strand DNA probe (Telo-probe) and centromere specific single strand DNA probe (Centro-probe) are added to the extracted genome DNA. Telo-probe and Centro-probe are labeled with Raman molecules 6-Carboxytetramethylrhodamine (TAMRA) and Methylene Blue (MB), respectively. They can hybridize specifically with telomeres or centromere. (3) After the two SERS probes have hybridized with genome DNA, magnetic capturing nanoparticles (NPs) are added to capture and remove the excess SERS probes. (4) Genome DNA hybridized with Telo-probe and Centro-probe is dropped onto a SERS substrate and subjected to SERS measurement. (5) The SERS spectra is collected and analyzed to get telomere length information. In the experiments, one normal cell (MRC5) and two cancer cell (HeLa and LNCaP) lines are used to test the feasibility of the proposed protocol. Telomere length variation in the proliferation of MRC5 cells is investigated as well as the influence of telomerase inhibitor on the telomere length of cancer cells.
Results
Successful fabrication of the SERS substrate is a prerequisite in SERS based telomere length measurement. The SERS substrate is fabricated by assembling silver nanoparticles (Ag NPs) onto glass slide through electrostatic interactions. First, Ag NPs are synthesized. They are negatively charged due to the citrate ligand. Then positively charged amino groups are introduced onto the surface of glass slide by modifying with APTMS. After soaking the amino-functionalized glass slide into Ag NPs solution, Ag NPs can self-assemble onto the glass slide via the electrostatic interaction between Ag NPs and amino groups. Figure 2 (a) shows the SEM image of the fabricated SERS substrate. As can be seen, Ag NPs are uniformly assembled in a quite large area. The average diameter of the Ag NPs is approximately 44 nm (see the supplementary Fig. S1). Average surface coverage percentage of Ag NPs is estimated to be 18.3% by analyzing SEM images with Adobe Photoshop CS6 software. Typical gap size between adjacent Ag NPs is about 23 nm as determined by measuring 350 gaps between Ag NPs in SEM images using ImageTool software (see the supplementary Fig. S2). Besides, about 14% of the Ag NPs are closely packed with each other (see the enlarged SEM image in Figure 2(b)), resulting in Ag NP clusters. These closely packed Ag NPs can produce a lot of “hot spots”, which help to better enhance SERS signals41. Figure 2(C) is the TEM image of Ag NPs. The inset in Figure 2(C) shows the photograph of a blank glass slide and a SERS substrate. Compared with the blank glass slide, the SERS substrate exhibits a more greenish color. The extinction spectra of the blank slide and SERS substrate are shown in Figure 2(d). The SERS substrate shows increased extinction coefficient compared with the blank slide. The changes in color and extinction spectra of the SERS substrate are caused by the assembled Ag NPs. Besides, an obvious extinction band peaked at around 379 nm is observed in the spectrum of the SERS substrate, which originates from the surface plasmon resonance (SPR) of Ag NPs. The SPR band of Ag NPs in aqueous solution peaks at 409 nm and undergoes a significant blue shift (30 nm) when the Ag NPs are assembled onto glass slide. This is due to the fact that Ag NPs in solution are surrounded by water while Ag NPs on glass slide are exposed to the air and the refractive index of air is smaller than that of water42,43,44. A comparison between the SERS enhancing ability of the SERS substrate and bare glass slide is conducted, 10 μM Telo-probe solution is casted onto the SERS substrate or glass slide and the SERS spectra are collected. The results are shown in the supplementary Fig. S3. Obviously, Ag NPs assembled SERS substrates provide strong SERS signals of Telo-probe (i.e. TAMRA), while no SERS signal is obtained from the blank glass slide, indicating that the SERS substrate can generate an excellent SERS enhancement. The above results prove that fabrication of the SERS substrate is successful.
After successful fabrication of the SERS substrate, the practicability of this protocol is tested. The working principle of the proposed method is demonstrated in Figure 1. In the experiments, two Raman molecules labeled single DNA strands are used as the SERS probes to distinguish telomeres and centromere. Telomere probe (Telo-probe: 5′TAMRA-CCCTAACCCTAACCCTAA-3′) is labeled with TAMRA, while centromere probe (Centro-probe: 5′MB-AAACTAGACAGAAGCAT-3′) is labeled with MB. Telo-probe is complementary to the G-rich strand of telomeres and Centro-probe is complementary to a specific repetitive sequence in human centromere. Once mixed with genome DNA, Telo-probe and Centro-probe can attach to the genome DNA through hybridization with telomeres and centromere. When the hybridization between SERS probes and genome DNA is finished, magnetic capturing NPs (Telo-capture and Centro-capture) are added to capture and remove excess SERS probes. Telo-capture and Centro-capture are obtained by attaching thiolated Telo-probe capturing strand (5′-SH(CH2)6-TTATAGGG-3′) or Centro-probe capturing strand (5′-SH(CH2)6-TTTAGCTTCTGTCTAGTTT-3′) onto gold coated magnetic nanobeads as described in the experimental section. Telo-probe capturing strand contains a mismatched base while Centro-probe capturing strand contains two mismatched bases (bold underlined bases). In this way, the binding affinity between SERS probes and genome DNA is larger than that between SERS probes and capturing strands. This ensures that Telo-capture and Centro-capture will only capture and remove excess unbound SERS probes, while the SERS probes already hybridized with genome DNA will remain attached. After removing the excess SERS probes by magnetic separation, the genome DNA attached with SERS probes is dropped on a SERS substrate for SERS spectra acquisition. With longer telomeres, more Telo-probes will attach to the genome DNA, resulting in a more stronger SERS signal of TAMRA. So SERS intensity can be used as the indicator of telomere length. While the telomere length varies from cell to cell, centromere length remains constant. Consequently, centromere length (or SERS signal of MB) can be used as an inner control for the calibration or normalization of telomere length, which can effectively eliminate inaccuracy caused by variation in genome DNA concentration45,46.
In the experiment, TAMRA and MB are selected as the Raman labels for Telo-probe and Centro-probe, respectively. Figure 3(a) is the SERS spectra of Telo-probe and Centro-probe obtained by directly casting the probe solutions onto a SERS substrate. All the observed SERS bands are typical for TAMRA and MB47,48. Specifically, the 628 cm−1, 1648 cm−1 peaks of TAMRA and 450 cm−1, 1622 cm−1 peaks of MB are clearly distinguishable from each other with no obvious overlapping. This allows robust discriminating between the SERS signals of Telo-probe and Centro-probe.
Next, telomere length measurement is conducted using genome DNA extracts of MRC5, HeLa and LNCaP cells as describe in the experimental section. The SERS spectra obtained are shown in Figure 3(b). In the blank control, genome DNA is replaced by PBS buffer solution. For genome DNA extracts, strong SERS signals are acquired. While for the blank control, negligible SERS signal is detected. This means that in blank control, with no genome DNA present, almost all the excess SERS probes are captured and removed by Telo-capture and Centro-capture, resulting in a very weak SERS signal which is probably caused by a few remanent SERS probes. On the contrary, with genome DNA present, SERS probes can hybridize and attach to the genome DNA, escape the subsequent magnetic separation and eventually produce obvious SERS signals. The above experimental results confirm that the proposed “hybridize, capture and SERS measurement” procedures work well. Worth mentioning is that since Telo-probe, Centro-probe and genome DNA are mixed together in a single tube, the final SERS spectra contains both the SERS signals of TAMRA and MB. Fortunately, owing to the well separated SERS peaks of TAMRA and MB (Figure 3(a)), distinguishing between the SERS signals of TAMRA and MB can be easily achieved.
After validating the feasibility of the SERS based protocol, telomere length measurement is performed using MRC5 cells. MRC5 is a normal cell line which lacks telomerase and ALT activity, so the telomeres of MRC5 undergo gradual shortening with increased cell division cycles, which renders MRC5 finite replicative capacity (42–46 population doublings, PD)49,50,51. Genome DNAs of MRC5 with different population doubling levels (PDL) are collected and subjected to the SERS based telomere length measurement assay. The results are shown in Figure 4. MRC5 (1) represents primary MRC5 cells, MRC5 (2) represents MRC5 cells with 20 PD, while MRC5 (3) represents MRC5 cells with 40 PD. As can be seen from Figure 4(a), the 1648 cm−1 peak intensity decreases with increased PDL. This is reasonable since the telomeres of MRC5 shorten with increased PDL, thus less Telo-probe is hybridized and attached to genome DNA, resulting in weaker SERS signal. However, it is inappropriate to simply use the absolute intensity of Telo-probe as an indicator of telomere length, because changes in genome DNA concentration can also affect the amount of hybridized SERS probes as well as the SERS intensity. Hence, centromere is used as the inner control for the calibration of the telomere length. To better describe telomere length, telomeres-to-centromere ratio (T/C ratio = ITAMRA-1648/IMB-1622) is introduced. Here, ITAMRA-1648 represents the SERS intensity of Telo-probe at 1648 cm−1, while IMB-1622 represents the SERS intensity of Centro-probe at 1622 cm−1. Gaussian peak fitting of the SERS spectra is performed in Origin 8.5 to get the intensities of the SERS peaks. The left pannel in Figure 4(b) shows the calculated T/C ratios obtained from the SERS spectra in Figure 4(a), which more vividly shows the gradual decrease in T/C ratio (i.e. telomere length).
(a) SERS spectra obtained using genome DNA extracts of MRC5 (1), MRC5(2) and MRC5 (3). (b) Calculated T/C ratios in SERS and FISH assays. MRC5 (1): primary cell, MRC5 (2): PDL = 20, MRC5 (3): PDL = 40. In SERS assay, 8 SERS measurements are performed for each sample and the error bars represent the standard deviation of the 8 measurements. In FISH assay, 30 cells are analyzed for each sample and the error bars represent the standard deviation of the 30 cells.
To examine the reliability of the above results, FISH assay is conducted using two fluorescent PNA probes, that is, Cy3 labeled telomere PNA probe (TelC-Cy3: N-CCCTAACCCTAACCCTAA-C) and FAM labeled centromere PNA probe (Cent-FAM: N-AAACTAGACAGAAGCAT-C). Cell nucleus is stained by NucRed® Live 647 (Invitrogen). Figure 5 is the fluorescence images obtained in FISH assay. As can be seen, telomeres and centromere are successfully stained with the fluorescent PNA probes. Here, T/C ratio = ITelC-Cy3/ICent-FAM is used to describe telomere length, in which ITelC-Cy3 is the fluorescence signal intensity of telomeres and ICent-FAM is the fluorescence signal intensity of centromere. The calculated T/C ratios are presented in the right panel of Figure 4(b). Comparing the T/C ratios in SERS and FISH assays, it can be found that the results in SERS assay corresponds well with FISH assay. Both of them show the decrease in telomere length with increased PDL. The difference in the value of T/C ratios between SERS and FISH assays may be caused by several reasons. First, in FISH assay, T/C ratio is calculated from the images of only a few cells. While in SERS based protocol, genome DNA is extracted from approximately 106 cells, so the T/C ratio is an average of an enormous amount of cells. Second, SERS spectra and fluorescence images are acquired with two independent detection systems, the difference in the sensitivity of the two systems may have an impact on T/C ratios. Third, DNA probes are used in the SERS protocol while PNA probes are used in FISH assay, the difference in binding affinity of DNA and PNA probes can also alter the T/C ratio. However, the difference in the T/C ratio values does not affect the evaluation of the reliability of the SERS based telomere length measurement protocol. It is noteworthy that the SERS based protocol produces relative telomere length (i.e. T/C ratio). Absolute telomere length can be obtained by correlating the T/C ratio with Southern blot, which is also the way how absolute telomere length is acquired in FISH and HPA22,29.
Fluorescence images obtained in FISH experiments.
(a) MRC5 (1), (b) MRC5 (2), (c) MRC5 (3). The first column is fluorescence images of the telomere, the second column is fluorescence images of the centromere, the third column is fluorescence images of the nucleus and the last column is merged images of the telomere, centromere and nucleus. For telomere the excitation wavelength is 543 nm and 585–615 nm lights are collected, for centromere the excitation wavelength is 488 nm and 500–530 nm lights are collected, while for nucleus the excitation wavelength is 633 nm and 670–700 nm lights are collected, respectively. The experimental parameters (i.e. laser power, scanning speed etc.) are kept the same.
Next, the SERS based telomere length measurement protocol is used to evaluate the telomerase inhibition effect of the drug molecule azidothymidine (AZT). AZT can be incorporated into genome DNA (since its molecular structure is very similar to thymidine) and subsequently inhibit the catalytic ability of the telomerase reverse transcriptase (TERT), which eventually inhibits telomerase based telomere elongation52,53,54. In the experiments, HeLa and LNCaP cells are used as the model telomerase positive cancer cells. They are first incubated with AZT (0.8 mM) for 2 weeks. Previously published work suggested that 0.8 mM AZT can induce observable telomere shortening54. Then the genome DNAs are collected and subjected to telomere length measurement. Cells incubated in the absence of AZT are used as the control. The results are shown in Figure 6. HeLa (1) and LNCaP (1) represent primary cells, HeLa (2) and LNCaP (2) represent cells incubated for 2 weeks in the absence of AZT, HeLa+AZT and LNCaP+AZT represent cells incubated with AZT for 2 weeks. For HeLa (2) and LNCaP (2), the calculated T/C ratios are comparable to HeLa (1) and LNCaP (1), indicating no telomere shortening has occurred during this time period. This is rational as telomerase in these cancer cells can maintain telomere length. While for HeLa+AZT and LNCaP+AZT, remarkable decrease in T/C ratios are observed compared with early PD cells and control cells. This means that with AZT present, the telomere elongation function of telomerase is well inhibited, resulting in telomere repeats loss, i.e. telomere length shortening. FISH assay is also performed and the results are shown in Figure 7 and Figure 6 (b, d). The fluorescence images and T/C ratios obtained in FISH also indicate shortened telomeres with AZT present. These results confirmed that AZT is an efficient telomerase inhibitor and the SERS based protocol is applicable in evaluating the inhibition effect of anti-cancer drugs.
(a) SERS spectra obtained using genome DNA extracts of HeLa (1), HeLa (2) and HeLa+AZT. (b) Calculated T/C ratios of HeLa cells in SERS and FISH assay. (c) SERS spectra obtained using genome DNA extracts of LNCaP (1), LNCaP (2) and LNCaP+AZT. (d) Calculated T/C ratios of LNCaP cells in SERS and FISH assay. In SERS assay, 8 SERS measurements are performed for each sample and the error bars represent the standard deviation of the 8 measurements. In FISH assay, 30 cells are analyzed for each sample and the error bars represent the standard deviation of the 30 cells.
Fluorescence images obtained in FISH experiments.
(a) HeLa (1), (b) HeLa+AZT, (c) LNCaP (1), (d) LNCaP+AZT. The first column is fluorescence images of the telomere, the second column is fluorescence images of the centromere, the third column is fluorescence images of the nucleus and the last column is merged images of the telomere, centromere and nucleus. Experimental parameters are kept the same to Figure 5.
Discussion
As can be seen from the SERS spectra in Figure 3, Figure 4 and Figure 6, the SERS peak at 450 cm−1 seem to be higher in HeLa and LNCaP compared with that in early PD MRC5 cells (i.e. MRC5 (1) and MRC5 (2)). As shown in Figure 3a, The Raman peak at 450 cm−1 is attributed to MB labeled Centro-probe. The fact that the 450 cm−1 peak in MRC5 is weaker than that in HeLa and LNCaP cells might be due to two reasons. First, in the experiments, the genome DNA extractions of HeLa and LNCaP have a higher concentration because these two cancer cells grow quite faster than MRC5 cells and more HeLa and LNCaP cells are used in genome DNA extraction than MRC5 cells. Higher genome DNA concentration means higher centromere concentration and thus stronger Centro-probe signal. Second, the SERS spectra of HeLa and LNCaP are dominated by the signal of Centro-probe (i.e. MB), while the SERS spectra of early PD MRC5 are dominated by the signal of Telo-probe (i.e. TAMRA). Consequently, telomeres-to-centromere ratios (T/C ratios) of HeLa and LNCaP cells are smaller than that of early PD MRC5 cells (Figure 4 and Figure 6). This is another reason why the SERS peaks of Centro-probe (450 cm−1, 1622 cm−1) in HeLa and LNCaP appeared to be higher than those of early PD MRC5 cells, while the SERS peaks of Telo-probe (628 cm−1, 1648 cm−1) in early PD MRC5 appeared to be higher than those of HeLa and LNCaP cells. However, it is not the absolute peak intensity but the intensity ratio between Telo-probe and Centro-probe that matters. Because variation in genome DNA concentration or Ag NPs density on the SERS substrate can cause obvious difference in the absolute SERS intensity. This is why centromere is used as the inner control for the calibration of the telomere length. Moreover, since T/C ratios of early PD MRC5 cells are larger than HeLa and LNCaP cells, this indicates that early PD MRC5 cells have longer or bigger telomeres than HeLa and LNCaP cells. This result is consistent with previously reported telomere lengths of the three cell lines55,56,57,58. Also can be seen from Figure 4 and Figure 6 is that FISH assay results in much larger error bars compared with SERS assay, this might be due to the fact that FISH only analyzes several cells while SERS protocol evaluates an enormous amount of cells. Consequently, SERS assay provides averaged results with smaller standard deviation. This means that compared with FISH, the presented SERS protocol can provide more accurate information on average telomere length. However, from another point of view, FISH can analyze single telomeres while the proposed SERS protocol is based on genome DNA extraction of a large number of cells. So even after correlating the T/C ratio of SERS assay with Southern blot, only the average value of the absolute telomere lengths can be obtained. The present SERS protocol can not provide individual telomere length of single cells. More effort is still needed to figure out a way to measure individual telomere length of single cells using SERS technique.
To test the reliability of the SERS based telomere length measurement method, genome DNA from three different batches of MRC5 cells with different PDL is tested and the results are shown in the supplementary Fig. S4. As can been seen, the T/C ratios of different batches of genome DNA extractions correspond well with each other, indicating that the novel SERS based technique has a good reproducibility and reliability.
Conclusions
A SERS based optical telomere length measurement protocol is presented and successfully realized. This protocol uses two different DNA SERS probes to simultaneously label telomeres and centromere, respectively. By measuring SERS signals and calculating the T/C ratios, information of telomere length can be obtained. FISH assay results confirm that the presented SERS protocol shows excellent reliability. The SERS based protocol can detect gradual telomere loss in MRC5 proliferation. It can also be used to evaluate the inhibition effect of telomerase inhibitors. This SERS based optical protocol does not require any PCR or electrophoresis procedures, which greatly simplifies operation. Besides, it is also cost-effective since no expensive PNA probe is needed. This easy-operation and reliable optical protocol is an attractive candidate for telomere length measurement and has a great potential in the investigation, diagnosis and therapeutics of telomeres/telomerase related diseases.
Methods
Materials
Silver nitrate (AgNO3), azidothymidine (AZT) and (3-aminopropyl)trimethoxysilane (APTMS) were purchased from Sigma-Aldrich. Absolute ethanol, sodium dodecyl sulphute (SDS), trisodium citrate dihydrate, NaCl, H2SO4 and H2O2 were purchased from Sinopharm Chemical Reagent Co., Ltd. Phosphate buffered saline (PBS, 10 mM, pH 7.4) and paraformaldehyde were purchased from Nanjing KeyGen Biotechnology Co., Ltd. Peptide nucleic acid (PNA) probes for FISH were purchased from Panagene (Korea). DNA probes were synthesized by Sangon Biotech (Shanghai) Co., Ltd. All the reagents were used as received. Deionized water (Millipore Milli-Q grade) with a resistivity of 18.2 MΩ/cm was used in all the experiments.
The sequences of PNA and DNA probes are as follows:
Cy3 labeled telomere PNA probe (TelC-Cy3): N-CCCTAACCCTAACCCTAA-C
FAM labeled centromere PNA probe (Cent-FAM): N-AAACTAGACAGAAGCAT-C
TAMRA labeled telomere DNA probe (Telo-probe): 5′TAMRA-CCCTAACCCTAACCCTAA-3′
MB labeled centromere DNA probe (Centro-probe): 5′MB-AAACTAGACAGAAGCAT-3′
Thiolated Telo-probe capturing strand: 5′-SH(CH2)6-TTATAGGG-3′
Thiolated Centro-probe capturing strand: 5′-SH(CH2)6-TTTAGCTTCTGTCTAGTTT-3′
Fabrication of the SERS substrate
The SERS substrate was obtained by assembling silver nanoparticles (Ag NPs) onto glass slides. First, Ag NPs were synthesized by reducing AgNO3 with trisodium citrate dihydrate according to previously published literature59. Glass slide was sonicated in a piranha solution (75% H2SO4 and 25% H2O2) for 1 h and then rinsed with copious amount of water. After drying with argon, the glass slide was immersed in absolute ethanol containing 2% APTMS for 12 h. Then the glass slide was washed with water, dried under argon and subsequently immersed in Ag NPs solution for 12 h. Finally, the glass slide with Ag NPs assembled on it was carefully taken out, washed with water and dried under argon. Thus the SERS substrate was obtained.
Fabrication of the magnetic capturing NPs
Gold coated magnetic nanobeads were synthesized according to previously published work60,61,62. To attach thiolated capturing strand, 300 μL (20 μM in PBS) of Telo-probe capturing strand or Centro-probe capturing strand was added to 300 μL of gold coated magnetic nanobeads. The mixture solution was sonicated for 10 s and shaken for 12 h. Next, 100 μL of the aging buffer (pH 7.4, 10 mM PBS, 0.6 M NaCl, 0.2% SDS) was added 10 times with an interval of 1 h. Finally, the capturing stand modified magnetic nanobeads were collected by magnetic separation. The sediments were dispersed in 150 μL of PBS and stored at 4°C before use. The Telo-probe capturing strand modified magnetic nanobeads were denoted as Telo-capture, while the Centro-probe capturing strand modified magnetic nanobeads were denoted as Centro-capture.
Cell culture and genome DNA extraction
Human cervical cancer cells (HeLa), human prostate cancer cells (LNCaP) and normal human embryonic lung fibroblasts (MRC5) were purchased from China Type Culture Collection. HeLa, MRC5 cells were cultured in DMEM and LNCaP cells were cultured in RPMI 1640. Cells were kept under standard cell culture condition (5% CO2, 37°C). Media were supplemented with 10% fetal bovine serum (GIBCO) and 1% penicillin–streptomycin (Nanjing KeyGen Biotech. Co., Ltd.). Genome DNA was extracted using a standard DNA extraction kit (KGA2750, Nanjing KeyGen Biotech. Co., Ltd.) according to manufacturer's instruction.
Telomere length measurement
For SERS based telomere length measurement, 25 μL of genome DNA extracts was mixed with 4 μL (10 μM in PBS) of Telo-probe and 4 μL (10 μM in PBS) of Centro-probe. The mixture was incubated on a Bio-Rad S1000 thermal cycler at 78°C for 10 min, 65°C for 5 min, 55°C for 5 min and 23°C for 12 h. Then 25 μL of Telo-capture and 25 μL of Centro-capture were added, the mixture solution was shaken for 3 h at room temperature. After that, the mixture solution was subjected to magnetic separation. Finally, the supernatant was dropped onto a SERS substrate for SERS measurement.
For FISH assay, cells were first seeded into 35 mm culture dish (glass bottom) and incubated for 24 h. Then the cells were fixed by 4% paraformaldehyde at room temperature for 20 min. After washing the culture dish twice with PBS, in-situ hybridization was performed according to manufacturer's instruction. When the FISH reaction was completed, the culture dish was dried under argon, sealed with antifade solution (Beyotime) and covered with cover slide. Finally, the culture dish was subjected to fluorescence measurement. Fluorescence images are analyzed by ImageJ software.
Instruments
Extinction spectra were measured by a Shimadzu UV-3600 PC spectrophotometer with quartz cuvettes of 1 cm path length. Transmission electron microscope (TEM) images were obtained with an FEI Tecnai G2T20 electron microscope operating at 200 kV. Scanning electron microscope (SEM) images were obtained by a Zeiss Ultra Plus electron microscope with 15 kV accelerating voltage. Fluorescence images were acquired by a confocal laser scanning microscopy (FV 1000, Olympus) with a 60× microscope objective. SERS spectra were recorded with a JOBIN YVON T64000 Raman system at 632.8 nm excitation using a 50× long working distance objective. The laser power was 13 mW at the sample position. The integrating time was 20 s.
References
de Lange, T. How telomeres solve the end-protection problem. Science 326, 948–952 (2009).
Palm, W. & de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 (2008).
Blackburn, E. H. Structure and function of telomeres. Nature 350, 569–573 (1991).
Blackburn, E. H. Switching and signaling at the telomere. Cell 106, 661–673 (2001).
Zakian, V. A. Telomeres - Beginning to understand the end. Science 270, 1601–1607 (1995).
Olovniko. Am. Theory of marginotomy - Incomplete copying of template margin in enzymic-synthesis of polynucleotides and biological significance of phenomenon. J. Theor. Biol. 41, 181–190 (1973).
Saldanha, S. N., Andrews, L. G. & Tollefsbol, T. O. Assessment of telomere length and factors that contribute to its stability. Eur. J. Biochem. 270, 389–403 (2003).
Blackburn, E. H. Telomere states and cell fates. Nature 408, 53–56 (2000).
Hayflick, L. & Moorhead, P. S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 (1961).
Mondello, C., Riboni, R., Casati, A., Nardo, T. & Nuzzo, F. Chromosomal instability and telomere length variations during the life span of human fibroblast clones. Exp. Cell Res. 236, 385–396 (1997).
Shay, J. W. & Bacchetti, S. A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791 (1997).
Collins, K. & Mitchell, J. R. Telomerase in the human organism. Oncogene 21, 564–579 (2002).
de Lange, T. Protection of mammalian telomeres. Oncogene 21, 532–540 (2002).
Feng, J. L. et al. The RNA Component of human telomerase. Science 269, 1236–1241 (1995).
Cesare, A. J. & Reddel, R. R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330 (2010).
Blasco, M. A. Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 6, 611–622 (2005).
Neidle, S. & Parkinson, G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 1, 383–393 (2002).
Oexle, K. Telomere length distribution and southern blot analysis. J. Theor. Biol. 190, 369–377 (1998).
Ishizuka, T., Xu, Y. & Komiyama, M. A chemistry-based method to detect individual telomere length at a single chromosome terminus. J. Am. Chem. Soc. 135, 14–17 (2013).
Cawthon, R. M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, 6 (2002).
Gil, M. E. & Coetzer, T. L. Real-time quantitative PCR of telomere length. Mol. Biotechnol. 27, 169–172 (2004).
Nakamura, Y. et al. Simple, rapid, quantitative and sensitive detection of telomere repeats in cell lysate by a hybridization protection assay. Clin. Chem. 45, 1718–1724 (1999).
Delara, J., Wydner, K. L., Hyland, K. M. & Ward, W. S. Fluorescent in-situ hybridization of the telomere repeat sequence in hamster sperm nuclear-structures. J. Cell. Biochem. 53, 213–221 (1993).
Lansdorp, P. M. et al. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5, 685–691 (1996).
Schneider-Stock, R. et al. On telomere shortening in soft-tissue tumors. J. Cancer Res. Clin. Oncol. 124, 165–171 (1998).
O'Callaghan, N. J., Dhillon, V. S., Thomas, P. & Fenech, M. A quantitative real-time PCR method for absolute telomere length. BioTechniques 44, 807–809 (2008).
Wang, F. et al. Robust measurement of telomere length in single cells. Proc. Natl. Acad. Sci. U. S. A. 110, E1906–E1912 (2013).
Poon, S. S. S., Martens, U. M., Ward, R. K. & Lansdorp, P. M. Telomere length measurements using digital fluorescence microscopy. Cytometry 36, 267–278 (1999).
O'Sullivan, J. N. et al. Telomere length assessment in tissue sections by quantitative FISH: Image analysis algorithms. Cytometry A 58A, 120–131 (2004).
Kalajzic, A. C., Vidacek, N. S., Huzak, M., Ivankovic, M. & Rubelj, I. Telomere Q-PNA-FISH - reliable results from stochastic signals. PLoS One 9, 10 (2014).
Fleischmann, M., Hendra, P. J. & McQuilla. Aj. Raman-spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 26, 163–166 (1974).
Campion, A. & Kambhampati, P. Surface-enhanced Raman scattering. Chem. Soc. Rev. 27, 241–250 (1998).
Camden, J. P., Dieringer, J. A., Zhao, J. & Van Duyne, R. P. Controlled plasmonic nanostructures for surface-enhanced spectroscopy and sensing. Acc. Chem. Res. 41, 1653–1661 (2008).
Xu, S. et al. Surface-enhanced Raman scattering studies on immunoassay. J. Biomed. Opt. 10, 031112–03111212 (2005).
Kneipp, K. et al. Single molecule detection using surface-enhanced Raman scattering (SERS). Phys. Rev. Lett. 78, 1667–1670 (1997).
Nie, S. M. & Emery, S. R. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science 275, 1102–1106 (1997).
Lim, D. K., Jeon, K. S., Kim, H. M., Nam, J. M. & Suh, Y. D. Nanogap-engineerable Raman-active nanodumbbells for single-molecule detection. Nat. Mater. 9, 60–67 (2010).
Luo, S.-C., Sivashanmugan, K., Liao, J.-D., Yao, C.-K. & Peng, H.-C. Nanofabricated SERS-active substrates for single-molecule to virus detection in vitro: A review. Biosens. Bioelectron. 61, 232–240 (2014).
Yamazoe, S. et al. Large-area surface-enhanced Raman spectroscopy imaging of brain ischemia by gold nanoparticles grown on random nanoarrays of transparent boehmite. ACS nano 8, 5622–5632 (2014).
Wang, Y., Yan, B. & Chen, L. SERS tags: Novel optical nanoprobes for bioanalysis. Chem. Rev. 113, 1391–1428 (2013).
Lu, Y., Liu, G. L. & Lee, L. P. High-density silver nanoparticle film with temperature-controllable interparticle spacing for a tunable surface enhanced Raman scattering substrate. Nano Letters 5, 5–9 (2004).
Kelly, K. L., Coronado, E., Zhao, L. L. & Schatz, G. C. The optical properties of metal nanoparticles: The influence of size, shape and dielectric environment. J. Phys. Chem. B 107, 668–677 (2003).
Mock, J. J., Barbic, M., Smith, D. R., Schultz, D. A. & Schultz, S. Shape effects in plasmon resonance of individual colloidal silver nanoparticles. J. Chem. Phys. 116, 6755–6759 (2002).
Lv, Y. et al. Antibiotic glass slide coated with silver nanoparticles and its antimicrobial capabilities. Polym. Adv. Technol. 19, 1455–1460 (2008).
Perner, S. et al. Quantifying telomere lengths of human individual chromosome arms by centromere-calibrated fluorescence in situ hybridization and digital imaging. Am. J. Pathol. 163, 1751–1756 (2003).
Vander Griend, D. J., Konishi, Y., De Marzo, A. M., Isaacs, J. T. & Meeker, A. K. Dual-label centromere and telomere FISH identifies human, rat and mouse cell contribution to multispecies recombinant urogenital sinus xenografts. Prostate 69, 1557–1564 (2009).
Yang, J. et al. Surface-enhanced Raman spectroscopy based quantitative bioassay on aptamer-functionalized nanopillars using large-area Raman mapping. Acs Nano 7, 5350–5359 (2013).
Guo, X. et al. Silver-gold core-shell nanoparticles containing methylene blue as SERS labels for probing and imaging of live cells. Microchim. Acta 178, 229–236 (2012).
Harley, C. B., Futcher, A. B. & Greider, C. W. Telomeres shorten during aging of human fibroblasts. Nature 345, 458–460 (1990).
Taylor, L. M. et al. Inactivation of p16(INK4 alpha), with retention of pRB and p53/p21(cip1) function, in human MRC5 fibroblasts that overcome a telomere-independent crisis during immortalization. J. Biol. Chem. 279, 43634–43645 (2004).
Allsopp, R. C. et al. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 89, 10114–10118 (1992).
Gan, Y. B. et al. Telomere maintenance in telomerase-positive human ovarian SKOV-3 cells cannot be retarded by complete inhibition of telomerase. FEBS Lett. 527, 10–14 (2002).
Ji, H. J. et al. Cyclic induction of senescence with intermittent AZT treatment accelerates both apoptosis and telomere loss. Breast Cancer Res. Treat. 93, 227–236 (2005).
Gomez, D. E., Armando, R. G. & Alonso, D. F. AZT as a telomerase inhibitor. Front. Oncol. 2, 113–113 (2012).
Von Zglinicki, T., Pilger, R. & Sitte, N. Accumulation of single-strand breaks is the major cause of telomere shortening in human fibroblasts. Free Radic. Biol. Med. 28, 64–74 (2000).
Sitte, N., Saretzki, G. & Von Zglinicki, T. Accelerated telomere shortening in flbroblasts after extended periods of confluency. Free Radic. Biol. Med. 24, 885–893 (1998).
Canela, A., Vera, E., Klatt, P. & Blasco, M. A. High-throughput telomere length quantification by FISH and its application to human population studies. Proc. Natl. Acad. Sci. U. S. A. 104, 5300–5305 (2007).
Zhang, B. et al. Telomere and microtubule targeting in treatment-sensitive and treatment-resistant human prostate cancer cells. Mol. Pharmacol. 82, 310–321 (2012).
Lee, P. C. & Meisel, D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J. Phys. Chem. 86, 3391–3395 (1982).
Zong, S. et al. Colorimetry and SERS dual-mode detection of telomerase activity: combining rapid screening with high sensitivity. Nanoscale 6, 1808–1816 (2014).
Deng, H. et al. Monodisperse magnetic single-crystal ferrite microspheres. Angew. Chem. Int. Ed. 44, 2782–2785 (2005).
Cong, H., Toftegaard, R., Arnbjerg, J. & Ogilby, P. R. Silica-coated gold nanorods with a gold overcoat: Controlling optical properties by controlling the dimensions of a gold−silica−gold layered nanoparticle. Langmuir 26, 4188–4195 (2009).
Acknowledgements
This work was supported by the National Key Basic Research Program of China (grant no. 2015CB352002), the Excellent Youth Foundation of Jiangsu Province (BK20140023), the Natural Science Foundation of China (NSFC) (Nos. 61275182, 61177033, 21104009), Science Foundation for The Excellent Youth Scholars of Southeast University, the Scientific Research Foundation of Graduate School of Southeast University (YBPY1310), the Scientific Innovation Research Foundation of College Graduate in Jiangsu Province (CXZZ12_0094) and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
S.Z., Z.W. and Y.C. designed the protocol. S.Z. carried out the experiments. Z.W. and Y.C. supported and advised the experiments. H.C. provided cells. S.Z., Z.W. and Y.C. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Zong, S., Wang, Z., Chen, H. et al. Assessing Telomere Length Using Surface Enhanced Raman Scattering. Sci Rep 4, 6977 (2014). https://doi.org/10.1038/srep06977
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06977
This article is cited by
-
Intracellular imaging and concurrent pH sensing of cancer-derived exosomes using surface-enhanced Raman scattering
Analytical and Bioanalytical Chemistry (2021)
-
Fabrication of Ordered Mullite Nanowhisker Array with Surface Enhanced Raman Scattering Effect
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.