Abstract
Turner syndrome is caused by complete or partial loss of the second sex chromosome and is characterized by spontaneous fetal loss in >90% of conceptions. Survivors possess an array of somatic and germline clinical characteristics. Induced pluripotent stem cells (iPSCs) offer an opportunity for insight into genetic requirements of the X chromosome linked to Turner syndrome. We derived iPSCs from Turner syndrome and control individuals and examined germ cell development as a function of X chromosome composition. We demonstrate that two X chromosomes are not necessary for reprogramming or maintenance of pluripotency and that there are minimal differences in gene expression, at the single cell level, linked to X chromosome aneuploidies. Formation of germ cells, as assessed in vivo through a murine xenotransplantation model, indicated that undifferentiated iPSCs, independent of X chromosome composition, are capable of forming germ-cell-like cells (GCLCs) in vivo. In combination with clinical data regarding infertility in women with X chromosome aneuploidies, results suggest that two intact X chromosomes are not required for human germ cell formation, qualitatively or quantitatively, but rather are likely to be required for maintenance of human germ cells to adulthood.
Similar content being viewed by others
Introduction
Turner syndrome occurs with complete or partial loss of the second sex chromosome (45,X) in 1–2% of all female conceptions. In more than 90% of cases, pregnancies are not carried to term1. Diverse somatic characteristics are associated with surviving Turner syndrome females, including short stature and cardiovascular abnormalities2,3. In addition, most Turner syndrome females are also infertile, establishing a link between the X chromosome and germ line formation and/or maintenance4,5. Only the lack of a second sex chromosome results in infertility as females with an additional X chromosome (Triple X syndrome) have normal fertility6.
Females have two X chromosomes, one active and one inactive in somatic cells. However, large regions of the silenced X chromosome, including the pseudoautosomal regions (PAR) and loci scattered across the chromosome, escape X chromosome inactivation (XCI)7. Thus, loss of one X chromosome in Turner syndrome females is hypothesized to lead to haploinsufficiency of genes that escape XCI, which may be required in two copies for normal development, including formation and/or maintenance of germ cells. For example, haploinsufficiency of SHOX, located in PAR1, is known to contribute to the short stature of Turner syndrome females8. Reproductive phenotypes of a murine model of Turner syndrome have been examined and although XO mice are fertile, they have approximately half the number of oocytes as XX mice postnatally and a shortened reproductive lifespan9,10,11. The species-specific difference in severity of germ cell defects has been linked to the ability of the single X chromosome in XO mouse germ cells to form non-homologous associations with an autosome or itself, while human XO germ cells are more likely lost early in meiosis before pairing has started12. Another hypothesis for the species differences may be linked to the fact that 15% of genes escape XCI in humans, in comparison to only a few in mouse7,13,14. Indeed, the majority of Turner syndrome females are infertile with greatly impaired germ cell development.
The differences in fertility phenotypes between mouse and human and the inaccessibility and difficulty of obtaining human germ cells during fetal development, suggested a need for alternative genetic tools to probe the genetic requirements of the X chromosome in human germ cell development. Induced pluripotent stem cells (iPSCs) offer a source of patient-specific stem cells that can be used to study germ cell development in vitro and in vivo15,16,17,18,19,20. Studies using mouse embryonic stem cells (mESCs) have revealed key transcription factors for germ cell specification21. Furthermore, the overexpression of these transcription factors can direct primed epiblast-like cells (EpiLCs), but not mESCs, to functional primordial germ cell (PGC)-like cells that contribute to spermatogenesis and offspring22. In other studies in vitro-derived germ cell-like cells were matured within the gonadal environment to provide evidence for germ line contribution through production of offspring23,24,25.
In the last two decades, methods of germ cell transplantation and xenotransplantation have been developed26. Murine seminiferous tubules can maintain autologous spermatogonial stem cells as well as those from different species, including humans; this suggests that many factors necessary to support and maintain proliferation and differentiation of human spermatogonial stem cells are conserved among species27,28. However, the fate of germ cells following xenotransplantation depends on evolutionary distance such that meiotic initiation and progression is not observed in transplanted spermatogonial stem cells if there is a large evolutionary distance between donor and host29. The duct and seminiferous tubule system of the male murine testis has been demonstrated to offer a permissive somatic environment for xenotransplantation of human male pluripotent stem cells from various genotypes to form germ-cell-like cells (GCLCs)19,20. The ovarian stroma does not offer a similar advantage for study due to inability to sequester the pluripotent stem cells within the germ cell niche resulting in teratoma production. Because of this limitation and given that male pluripotent cells produced early GCLCs19,20, we investigated the ability of normal female and X aneuploid pluripotent stem cells to form early GCLCs within the murine seminiferous tubules.
Results
Reprogramming of fibroblasts from Turner syndrome and control females
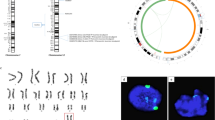
We obtained fibroblasts from a 30-year-old female (Control with normal fertility) and a 32-year-old female with premature ovarian failure (POF) as well as four Turner syndrome female fibroblast lines — TSN from a 1-day-old, TSF from an 18-week-old, TSC1 from a 3.5-year-old and TSC2 from an 8-year-old (Supplemental Table S1). Spectral Karyotyping (SKY) analysis confirmed the presence of two X chromosomes in the Control and POF fibroblasts, whereas all Turner fibroblast lines had only one X chromosome (Fig. 1A). In addition to SKY, we used a whole genome single nucleotide polymorphism (SNP) array to examine the X chromosome at a higher resolution. Array analysis demonstrated that Control fibroblasts had biallelic X-linked SNPs, while Turner fibroblasts had monoallelic SNPs from the single X chromosome, except for TSC1, which had biallelic SNP expression from the centric portion (p11.1 – q13.1; Fig. 1B).
Reprogramming of Control, POF and Turner syndrome fibroblasts to iPSCs.
(A) Spectral karyotype of Control, POF and Turner syndrome (TSC1, TSC2, TSN and TSF) fibroblasts. White box indicates two X chromosomes in Control and POF fibroblasts and only one X chromosome in all Turner syndrome fibroblasts. (B) X chromosome deletion map from genome-wide human SNP array for fibroblast samples. Red shaded box indicates region of monoallelic SNPs, indicating deletion of second X chromosome in Turner syndrome fibroblasts. Arrowhead marks centric portion of TSC1 with biallelic SNPs. (C) Phase contrast image of one iPSC subclone (C = subclone) colony, grown on MEFS or matrigel, derived from Control, POF and Turner syndrome fibroblasts along with their spectral karyotype. White box indicates two X chromosomes in Control and POF iPSC subclones and only one X chromosome in all Turner syndrome iPSC subclones. (D) Immunofluorescence for cell surface markers TRA-1-60, TRA-1-80 and SSEA4 (green) along with nuclear marker OCT4 (red) marking pluripotent iPSC subclones. Cell nuclei were costained with DAPI (blue). Scale bar, 150 μm. (E) Immunofluorescence for markers of the three germ layers after spontaneous differentiation of iPSC subclones, demonstrating cells expressing α Feto Protein (endoderm), Smooth Muscle Actin (mesoderm) and βIII Tubulin (ectoderm) (green). Cell nuclei were costained with DAPI (blue). Scale bar, 150 μm. (F) In vitro (teratoma) differentiation of subclones from all iPSC lines with evidence of all three germ layers, gut epithelium (endoderm), cartilage and smooth muscle (mesoderm) and neural rosettes and pigmented epithelium (ectoderm).
We reprogrammed fibroblasts through either retroviral transduction with four transcription factors separately (OCT4, SOX2, KLF4 and CMYC) or lentiviral transduction of the STEMCCA cassette carrying all reprogramming factors in a polycistronic vector (Supplemental Fig. S1A)30. We observed iPSC colonies after 11–32 days post transduction (Fig. 1C and Supplemental Fig. S1B). In one case, with TSC1 fibroblasts, reprogramming required addition of valproic acid (VPA). VPA is a histone deacetylase that was previously shown to increase the efficiency of reprogramming primary human fibroblasts to iPSCs31. We confirmed that all iPSC lines and subclones demonstrated the same karyotype as the original fibroblast lines (Fig. 1C and Supplemental Fig. S1B). Moreover, all iPSC subclones expressed the cell surface pluripotency markers, TRA-1-60, TRA-1-81 and SSEA432 and the nuclear pluripotency marker OCT4 (Fig. 1D and Supplemental Fig. S1C). We also demonstrated the formation of the three germ layers after embryoid body spontaneous differentiation, showing that cells formed endoderm (α-fetoprotein), mesoderm (Smooth Muscle Actin) and ectoderm (βIII Tubulin; Fig. 1E and Supplemental Fig. S1D). When iPSCs were injected either subcutaneously or under the kidney capsule of female immunodeficient mice, all iPSC lines formed teratomas with structures representative of the three primary germ layers (Fig. 1F). This indicated that X chromosome aneuploidy does not affect reprogramming to pluripotency or differentiation into the three primary germ layers, similar to a previous report of iPSC-derived teratoma formation with Turner lines33.
Single cell expression analysis of pluripotency and X-linked genes in control and X aneuploidy iPSCs
In humans, it is estimated that up to 15% of genes escape XCI, in comparison to only a few genes in mouse7. This difference may explain mild phenotypes seen in XO mice13,14. The majority of genes that escape XCI are located in the recombining pseudoautosomal region 1 and 2 (PAR1 and PAR2) at the tips of the X chromosome or have a Y chromosome homolog7,34. We examined whether genes that escape XCI are expressed at a lower level in Turner syndrome iPSCs relative to H9 (46,XX) human embryonic stem cells (hESCs); for this purpose, we analyzed single cells of all iPSC subclones, including a Triple X iPSC line with an additional X chromosome.
To measure gene expression in single cells, we sorted hESCs and iPSCs for single cells positive for SSEA4 and TRA-1-60, two antigens that characterize pluripotent stem cells32 (Fig. 2A). The percentage of double-positive cells ranged from 73.5–97% and all single cells were sorted from a >95% pure double-positive population (Supplemental Fig. S2A). We first assessed pluripotency gene expression in subclones of all iPSCs and compared them to H9 hESCs. We identified a few subclones with a significant difference in expression of multiple pluripotency genes (p-value < 0.01); however, this was not consistent across all subclones of an iPSC line or across more than two pluripotency genes (Fig. 2B). Additionally, we identified low or no expression of common germ layer genes, except for KDR, a common mesodermal associated gene; however, KDR was also highly expressed in H9 hESCs (Supplemental Fig. S2B). We also identified single cells positive for germ cell related genes including CKIT and GDF3 (Supplemental Fig. S2C). These results indicate small subclonal differences in expression of pluripotency, germ layer or germ cell-related genes independent of X chromosome composition.
Single cell analysis of pluripotency genes and pseudoautosomal regions and genes that escape XCI in pluripotent stem cells.
(A) Schematic of sorting method used to identify and isolate single pluripotent cells for gene expression analysis. (B–E) Violin plot representation of Log2Ex values (fold change above background) of single cells for (B) pluripotency genes in fibroblasts, hESC or iPSC subclones (C = subclone), (C) XIST, (D) genes on PAR1 and PAR2 and (E) genes that escape XCI genes on the X chromosome in hESC and iPSC subclones. Asterisk indicates a significant difference in expression (p-value < 0.01) when compared to H9 hESCs. Color-coded subclone information is provided.
The long non-coding RNA, XIST, marks the inactive X chromosome35,36. By using RNA FISH for XIST, we found Control iPSC subclones 7 and 8 displayed mosaic marks of XIST, while XIST was not detected in clone 1 (Supplemental Fig. S3A). We confirmed this finding by analyzing Histone H3 Lysine 27 trimethylation (H3K27me3), which also marks the inactive X chromosome (Supplemental Fig. S3B)37,38. Triple X iPSCs contained multiple cells with two XIST and H3K27me3 foci, indicating the silencing of the second and extra X chromosome (Supplemental Fig. S3A and S3B). We then analyzed all iPSC subclones and Control and POF fibroblasts for XIST expression at a single cell resolution. Control and POF fibroblasts had expression of XIST in 100% of single cells (Fig. 2C) whereas single pluripotent cells from a male (XY) iPSC line did not demonstrate any XIST expression as expected. H9 hESC, subclones from Control and POF iPSCs demonstrated mosaic expression of XIST. However, Triple X iPSCs expressed high levels of XIST in 100% of single cells. The majority of subclones from all Turner syndrome iPSC lines did not express XIST, however each line did have few cells that did express low levels of XIST (Fig. 2C). These single cell reports support Mekhoubad et al. and Kim et al. which demonstrate X chromosome erosion in early and late passage female iPSCs39,40. Because of the mosaicism of XIST in female XX pluripotent cell lines, we focused our attention on genes that escape XCI and are likely not to be influenced by the presence or absence of XIST.
Next, we examined X-linked genes for differences in expression in subclones of all Turner syndrome iPSC lines by analyzing nine genes found within PAR1 and one within PAR2. First, most genes within the PAR regions had low expression within single cells, with Log2Ex expression values (fold change above background) ranging from 1.10 to 9.24, or were undetectable in many single cells (Fig. 2C). Both subclones of TSC1 iPSCs, in addition to TSC2 iPSC subclone 2 and TSF iPSC subclone 9 had significantly lower expression of CD99 (p-value < 0.01), but not all Turner syndrome iPSC subclones had lower expression of CD99 than H9 hESCs. Six other PAR genes had subclones with significant differences in Log2Ex values compared to H9 hESCs (p-value <0.01), but this was not seen amongst multiple subclones of one iPSC line.
We also analyzed genes that are found throughout the X chromosome but have demonstrated escape from XCI7,34. We first observed that the Log2Ex values from this set of X-linked genes was higher than those found within PAR regions, with an average Log2Ex value of 4.90 to 16.99 (Fig. 2D). Again, we found individual clones that did have significant differences (p-value <0.01) in Log2Ex values for 11 of the 12 escape XCI genes in comparison to H9 hESCs. However, only TSC2 iPSCs had both subclones with significantly lower expression (p-value < 0.01) than H9 hESCs when analyzed for ZFX transcript. One clone from TSC1, TSF and TSN iPSCs also had lower expression of ZFX transcript (Fig. 2D). We identified significantly higher expression of PRKX, NLGN4X, USP9X and RBMX within Triple X iPSCs relative to to H9 hESCs. Overall, single cell expression analysis revealed subclonal variations in PAR and escape XCI X-linked genes in TSC iPSCs in comparison to H9 hESCs. However, none of the genes analyzed demonstrated reduced expression levels across all subclones of the four TSC iPSC lines, indicating X-linked expression levels were independent of X chromosome number.
Single cell analysis of in vitro differentiation of control and X chromosome aneuploid pluripotent cells with BMP4/8 and retinoic acid
To compare in vitro PGC differentiation across genotypes, we used a VASA:GFP reporter, which has previously been used to isolate primordial germ cells from both female and male hESCs and iPSCs15,16,17,20, to identify single GFP+ cells daily throughout a seven-day germ cell differentiation with BMP4/8 and retinoic acid (RA; Fig. 3A and Supplemental Fig. S4A)16,20,41,42. At day 0 (undifferentiated hESC or iPSC) and throughout germ cell differentiation, we detected a population of GFP+ cells across all lines, while non-transfected cells sorted at day 0,1 and 7 did not contain GFP+ cells (Supplemental Fig. S4B). We analyzed the expression of a panel of 15 germ cell-related genes within single cells throughout the seven-day directed differentiation of H9 hESC, Control, TSC1 and Triple X iPSCs (Fig. 3B). The low percentage of VASA:GFP+ cells obtained throughout differentiation, the fact that VASA was found in less than half of GFP+ single cells and expression of the additional 14 germ cell-related genes was not enriched throughout the directed differentiation led us to examine the ability of undifferentiated cells to form PGCLCs through xenotransplantation.
Single cell in vitro differentiation analysis in pluripotent stem cells using VASA:GFP reporter.
(A) Schematic of in vitro differentiation of VASA:GFP transfected pluripotent stem cells across genotypes for seven days with BMP 4/8 and RA. GFP+ cells were sorted and then purity sorted each day for single cell qRT-PCR analysis. (B) Heatmap of single GFP+ cells from h9 hESC, Control iPSCs, TSC1 iPSCs and Triple X iPSCs throughout the seven day directed differentiation. Global Log2Ex values (fold change above background) of germ cell genes is shown with grey indicating no detectable expression.
RNA-Seq analysis of GFP+ and GFP- populations after in vitro directed differentiation
To confirm single cell gene expression data, we differentiated Control, TSC1,Triple X iPSCs and H9 hESCs with BMP4/8 and RA for seven days and performed whole transcriptome analysis on GFP+ and GFP- populations. While all pluripotent cells, independent of X chromosome number, showed similar global gene expression patterns, the differentiated cells showed distinct gene expression patterns (Fig. 4 A & B). However, GFP+ and GFP- populations from all the lines analyzed had similar gene expression profiles (Fig. 4A & B). Although the differentiated Control lines clustered separately from the other differentiated populations, this result was not based on chromosomal composition, as H9 GFP+ and GFP- populations clustered closely with TSC1 and Triple X differentiated populations. In agreement with single cell analysis of pluripotent cells, we saw minimal differences in gene expression levels between H9 hESC, Control, TSC1 and Triple X iPSCs regardless of the position on the X chromosome (Fig. 4C). After directed differentiation, there were minimal expression differences for genes located along the length of the X chromosome between all lines analyzed, independent of X chromosome composition (Fig. 4D).
RNA-Seq analysis of in vitro directed differentiation of pluripotent stem cells using the VASA:GFP reporter.
(A) Multi-dimensional scaling (MDS) plot of RNA-seq samples. Similarities of gene expression patterns were calculated and mapped for H9 hESCs, Control iPSCs (subclones 7 and 8), TSC1 iPSCs (subclones 1 and 2) and Triple X iPSCs as well as day 7 differentiated H9 cells sorted for GFP (H9 GFP- and H9 GFP+), day 7 Control subclone 8 cells sorted for GFP (Control GFP- and Control GFP+) day 7 differentiated TSC1 subclone 2 cells sorted for GFP (TSC1 GFP- and TSC1 GFP+) and day 7 differentiated GFP+ Triple X cells (Triple X GFP+). (B). Hierarchical clustering of X aneuploid samples, pairwise scatterplots of gene expression levels and the similarity matrix. The similarity matrix is colored from yellow to red, from the most similar to the most different sample comparisons. C) X chromosome scanning of pluripotent cells. The length of X chromosome was scanned in 10,000 bp windows. The reads mapping to each bin were normalized for the number of loci it maps to and the fraction of the mapped length that overlaps the bin. The reads from clones of control and TSC1 iPSCs were averaged. D) X chromosome scanning of differentiated cells. The reads mapping across the X chromosome was calculated as above. The GFP+ and GFP- reads were averaged for H9, Control and TSC1 differentiated cells.
To verify single cell results, we examined RNA-Seq data for some germ cell-related genes probed using qRT-PCR. The GFP+ population contained more reads for VASA but the difference was not significant when compared to the GFP- population (Supplemental Fig. S5A) and upon closer analysis of RNA-Seq data, many reads mapped to 5′ and 3′ untranscribed regions (UTR), indicating that the mapped reads came from the integrated VASA:GFP reporter, rather then endogenous VASA transcript. Only one germ cell related gene, GFRA1, was upregulated in the differentiated populations in comparison to pluripotent stem cells (Supplemental Fig. S5B). Many germ cell-related genes were expressed in pluripotent cells and their expression was downregulated upon differentiation in both GFP+ and GFP- cells (SALL4, OCT4, PRDM14, CKIT, GDF3, DAZL; Supplemental Fig. S5C). Moreover, germ cell-related gene PUM1 expression remained constant between undifferentiated and differentiated cells (Supplemental Fig. S5D). Due to the expression of multiple germ cell related genes in undifferentiated hESCs and iPSCs, including PRDM14, a pivotal gene required to transform mESCs to germ cell competent cells following transplantation24,43 and the low efficiency of in vitro PGC differentiation, we investigated alternative in vivo methods for germ cell formation from pluripotent stem cells.
In vivo germ cell formation by murine xenotransplantation of control and X chromosome aneuploid pluripotent stem cells
Recently, it was shown that the xenotransplantation of human male pluripotent stem cells into the seminiferous tubules of busulfan-treated recipient murine testes promoted the in vivo differentiation of these pluripotent stem cells into GCLCs19,20. Because of the evolutionary distance between mouse and human, it was expected that the development of cells would be limited to the formation of early stage GCLCs29. We hypothesized that female pluripotent stem cells transplanted within the male gonadal ridge would be directed to form early GCLCs, but that the cells derived from Turner syndrome patients may not be maintained, thus, leading to infertility phenotype in humans. We assessed the possibility of using this xenotransplantation assay as an experimental system to test the ability of X chromosome aneuploid and female pluripotent stem cells to form early GCLCs in vivo.
A total of six female pluripotent stem cell lines — H9 hESC, Control (subclone 7 and 8), TSC1 (subclone 1 and 2) and Triple X iPSCs — were transplanted into the seminiferous tubules of recipient testes (52 in total) of immunodeficient nude mice whose endogenous spermatogonial stem cell population has been disrupted by busulfan treatment (Fig. 5A and Supplemental Fig. S6A). The recipient mice were sacrificed eight-weeks post-transplantation and testis were weighed and fixed for serial sectioning for further analysis (Supplemental Table S4). The weight of testis xenografts at the time of harvesting and analysis of H&E staining of testis cross sections indicated that all pluripotent lines produced at least one sample with teratoma-like proliferation outside of the seminiferous tubules (Supplemental Fig. 6B and C). Teratoma formation was most marked in TSC1 subclone 2 injected testis, with four out of six testis weighing above 150 mg and structures indicating teratoma formation in serial sectioned testis stained with H&E (Supplemental Fig. B and C). However, reinjection of TSC1 subclone 2 (4246L & R, 4247L & R and 4248L & R) and TSC1 subclone 1 did not produce teratoma-like structures, suggesting that teratoma formation was not specific to the patient from which the cells were derived and could be due to the particular injection. As teratoma-like proliferation occurred outside the seminiferous tubules, these results emphasize the importance of the somatic environment upon pluripotent stem cell differentiation and cells that expand inside the seminiferous tubules and rupture the tubule receive signals to form teratoma-like structures rather than GCLCs.
In vivo germ cell formation of control and X chromosome aneuploid pluripotent stem cells within murine seminiferous tubules.
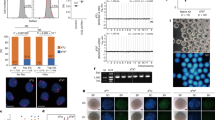
(A) Schematic of xenotransplantation procedure. Control and X chromosome aneuploid pluripotent stem cells were injected into the seminiferous tubules, via the efferent ducts of busulfan treated, immune deficient mice. (B) Histological cross sections from Human fetal testis (20 weeks), uninjected mouse testis, male control iPSCs, H9 hESCs, Control iPSCs, TSC1 iPSCs and Triple X iPSC-injected murine testis analyzed by immunohistochemistry using human specific antibody, NUMA and germ cell marker VASA antibody. All images include: i) single channel for NUMA (green); ii) VASA (red) co-stained with nuclear DAPI (blue); iii) merge of NUMA/VASA/DAPI. Scale bar, 40 um. White dashed line indicates border of seminiferous tubule. White box within merge demonstrates magnified region (Scale bar, 10 um) showing injected samples with human germ cells with colocalization of NUMA and VASA in murine seminiferous tubules (iv).
Next, we used immunostaining to further examine serial sections from injected mouse testis for human donor cells. To detect human cells, we used the human-specific antibody, NUMA20,44,45. NUMA staining was detected in all cells within a cross-section of a 20-week human fetal testis; in contrast, NUMA antibodies had no reactivity in cells from the un-injected mouse testis (Fig 5B). Male XY iPSC injected testis cross sections contained GCLCs as previously described that are readily identified as NUMA positive19 with cytoplasmic expression of VASA, a germ cell specific marker (Fig. 5B). We observed cells with similar morphology and positive staining for both NUMA and VASA proteins in the seminiferous tubules of testis that were injected with H9 hESC and Control iPSC as well as iPSCs with X chromosome aneuploidies (Fig. 5B). Thus, the xenotransplantation assay results demonstrated that H9 hESCs, Control, TSC1 and Triple X iPSCs all form GCLCs within mouse seminiferous tubules. We observed that the female pluripotent stem cell lines showed varying potential to form NUMA/VASA positive cells within the seminiferous tubules after xenotransplantation. All H9 hESC (4/4) and Triple X iPSCs (5/5) xenotransplants analyzed contained NUMA/VASA positive cells. Nine out of 15 testis with Control iPSCs contained double positive cells (60%) and TSC1 iPSCs had 12 out of 18 (66.7%) testis containing NUMA/VASA positive cells (Supplemental Fig. S7A and S7B). To further quantify our immunohistochemistry analysis across genotypes, we first counted the number of tubules that contained NUMA+/VASA+ cells to determine the percentage of total tubules that had early GCLC formation (Fig. 5C and Supplemental Fig. S7C). We observed a wide range of in the percentage of tubules with NUMA+/VASA+ cells, ranging from 2.59% up to 44.95% (Supplemental Table S4). No significant difference was observed amongst any of the xenotransplanted pluripotent stem cell lines, even when control female and X aneuploid pluripotent lines were compared to male hESC and iPSCs (Fig. 5C). Next, we counted the number of NUMA+/VASA+ cells within a positive tubule (Fig. 5D and Supplemental Fig. S7D). Only two transplants, H9 hESC and Triple X iPSCs, showed a significant difference (p-value = 0.017) in the number of NUMA+/VASA+ cells per positive tubule. Results indicate that the formation and specification of GCLCs within the in vivo xenotransplantation model are independent of sex chromosome number.
Human germ cells express STELLA, DAZL and the methylation mark H3K27me3
We extended our analysis of sections from H9 hESC, Control, TSC1 and Triple X iPSC xenotransplants to additional germ cell-related genes. Previous reports have described cKIT and OCT4 as markers of pre-gonadal PGCs followed by expression of VASA and DAZL in PGCs that may or may not continue to express OCT446,47. These cells are also enriched for the methylation mark, H3K27me3. Further maturation of germ cells results in depletion of H3K27me346. Reports have also described the importance of STELLA in early human germ cell formation48. All xenotransplant results were compared to cross-sections of a 20-week old human fetal testis and un-injected mouse testis section. Human fetal testis, which typically contain gonocytes and undifferentiated spermatogonia, contained cells positive for OCT4, CKIT and H3K27me3 proteins within the seminiferous tubules (Fig. 6A–E). Uninjected mouse testis contained cells positive for DAZL and H3K27me3 (Fig. 6B–E).
Analysis of additional germ cell related markers in in vivo derived germ cells.
(A–G) Immunofluorescence images of cross-sectioned Human fetal testis (20 week), mouse uninjected testis and xenografts from mouse testis injected with H9 hESC, Control iPSC, Triple X iPSC and TSC1 iPSCs. Positive NUMA (green) or STELLA (red) staining indicated human derived cells. Cross-sections were analyzed for (A) OCT4 (red), (B) DAZL (red), (C) STELLA (red), (D) CKIT (green) and (E) H3K27me3 (green). White arrow indicates human derived cells, marked with either NUMA or STELLA, that are positive for additional markers. Scale bar, 25 um.
We found no evidence for human-derived cells that expressed OCT4 in any of the transplanted testes (Fig. 6C), indicating that the cells observed post-xenotransplantation are not simply hESCs or iPSCs. We identified few NUMA+ cells that were also DAZL+ for each line transplanted (Fig. 6D). In contrast, all NUMA+ cells within the xenotransplants were also STELLA+ (Fig. 6E). Because of this finding, we used STELLA is a marker of human-derived cells within xenotransplants and co-stained with cKIT and observed that STELLA+ cells were not CKIT+. Lastly, we analyzed STELLA+ human-derived cells within the xenotransplants for presence of H3K27me3 and found that all human derived cells were positive for H3K27me3 (Fig. 6G). Taken together, results indicate that human cells that engrafted within the seminiferous tubules of recipient mouse testis expressed VASA, STELLA and H3K27me3, with only a small subset expressing DAZL; these results were consistent across all genotypes further indicating that the human GCLCs are likely at a gonocyte stage of development. Results demonstrate that the formation of GCLCs is independent of X chromosome composition, as Turner and Triple X iPSCs formed GCLCs comparably to Control pluripotent cells.
Discussion
To develop a model for Turner syndrome, we generated four Turner syndrome iPSC lines in addition to a POF and Control female iPSC line. We observed that all iPSC lines, independent of genotype, were capable of differentiating in vitro and in vivo into cell types representative of all three germ layers and the germ cell lineage. This demonstrates that two X chromosomes are not necessary for reprogramming or formation of somatic and germ cell lineages and is in agreement with a previous study that demonstrated that Turner Syndrome iPSCs differentiated into specific lineages indistinguishably from those produced from hESC and control iPSCs33. Moreover, our results indicate that the expression of XIST RNA is mosaic even within a single subclone. Only a small group of escape-XCI X-linked genes were expressed more highly in Triple X iPSCs (PRKX, NLGN4X, USP9X and RBMX), but there were few differences in X-linked gene expression at the single cell level compared to the control and X chromosome aneuploidy iPSCs to female H9 XX hESCs.
Investigations of germ cell differentiation in vitro indicated that the greatest similarity of cells, to germ cells, was observed on Day 0 (undifferentiated) indicative of inefficient differentiation. Thus, we used murine xenotransplantation to further examine germ cell development. Previous studies in the mouse used epiblast-like cells to explore formation of germ cells, also relying upon transplantation21,22,23,24,43. Thus, it is clear that transplantation of cells with germ cell differentiation potential into the correct somatic environment is able to promote germ cell development. Because multiple germ cell-related genes, especially PRDM14, are expressed in control female and X chromosome aneuploidy iPSCs prior to differentiation, we hypothesized that xenotransplantation of these cells into murine seminiferous tubules would enable germ cell development in vivo. Xenotransplantation was previously demonstrated to promote formation of GCLCs from male iPSCs including iPSCs derived from azoospermic men19,20; similarly, there is evidence that mouse PGCs have the potential to develop and progress through spermatogenesis or oogenesis49. Nonetheless, while mouse XX germ cells can enter spermatogenesis, they do not progress through meiosis, indicating the necessity of Y-specific transcription for further spermatocyte maturation50,51.
When XX human female pluripotent cells were transplanted into murine testes, the seminiferous tubules provided an environment to promote GCLC differentiation as anticipated. In addition, we also demonstrate formation of germ cells from Turner syndrome and Triple X iPSCs. Human-derived GCLCs migrate to the basement membrane within the seminiferous tubules and express the germ cell markers VASA, STELLA and DAZL and maintain the H3K27me3 mark suggesting the in vivo differentiated GCLCs are early in development and likely gonocytes. In addition Control female and Turner syndrome pluripotent stem cell transplants were quantitatively similar in terms of NUMA+/VASA+ tubules and NUMA+/VASA+ cells per positive tubules. Only Triple X xenotransplants contained fewer NUMA+/VASA+ cells per positive tubule when compared to H9 hESCs. These results indicate that the loss of an X chromosome does not affect germ cell formation, but suggest that an additional X chromosome may lead to decreased early GCLC formation within the seminiferous tubule environment which normally promotes XY germ cell formation.
In vivo derivation of GCLCs from patient-specific iPSCs opens the door to further studies on genetic mechanisms underlying female infertility. Human Control or X aneuploid derived GCLCs can be isolated for further use in single cell studies of X-linked gene expression. Indeed, here our single cell studies focused on genes that escape XCI. However, there are two stages of development where both X chromosomes are active -- in the cleavage stages of preimplantation embryo development and in female primordial germ cells. If Turner syndrome females are able to produce germ cells to the stage of X chromosome reactivation, then it is possible that haploinsufficiency of genes found throughout the entirety of the X chromosome may lead to germ cell loss. If similar xenotransplantation studies are to be accomplished with ovarian stroma, alternative methods are required to limit the dispersion of pluripotent cells while maintaining signals from the ovarian stroma. Previous studies have used aggregation of pluripotent cells with fetal or neonatal ovarian somatic cells prior to transplanting them under the mouse ovarian bursa24. Future studies with aggregation may allow examination of differences in later stages of development, on a female somatic background.
Overall, we demonstrate that undifferentiated iPSCs have expression of multiple germ cell associated genes and upon directed in vitro differentiation, this expression is lost and most cells are pushed to a somatic fate. Via xenotransplantation, we demonstrate that all pluripotent lines were capable of forming GCLCs, independent of X chromosome composition. These results indicate that infertility in Turner syndrome females is unlikely to be linked to defects in formation of germ cells in Turner syndrome women, but rather is likely to be linked to defective maintenance and/or further differentiation of germ cells.
Methods
Retroviral and lentiviral production, transduction and iPSC generation
Retroviral and lentiviral production was conducted as described previously30,52. Procedures for production of iPSCs were as described previously52,53. Briefly, one day pre-transduction, cells were plated at 7.5 × 105 cells/well of a six well plate. Retroviral supernatants (OCT4, SOX2, KLF4, CMYC) or lentiviral (STEMCCA) were mixed at a 10× concentration in fresh MEF media up to 1 ml and supplemented with polybrene (Sigma-Aldrich) at 8 ng/ml and added to fibroblasts for 24 hours. Two rounds of transduction were completed for retroviral transduction of fibroblasts and one round for lentiviral transduction. After transduction, cells were washed with PBS and fresh MEF media was added. MEF media with 0.5 mM Valproic acid (Sigma-Aldrich) was added to TSC1 fibroblasts. Five days post-transduction, cells were passaged to irradiated MEF feeders and switched to hESC media until colony formation. Upon visual detection of colonies, colonies were manually passaged and transferred to new irradiated MEF feeders. All iPSC and hESC lines were grown in 20% atmospheric oxygen at 37°C.
Single Cell qRT-PCR and Analysis
Single cell analysis was conducted using the BioMark HD System and the 96.96 dynamic array (Fluidigm). Individual cells in 2× reaction mix were preamplified for specific targets by the addition of SuperScriptIII RT PlatinumTaq Mix (Life Technologies), 200 nM of pooled primers (Supplemental Table S2) and DNA suspension buffer (Teknova). Lysis and specific target amplification were performed at 50°C for 15 min., followed by 95°C for 2 min. Sequence target amplification (STA) was performed by 18 cycles of 95°C denaturation for 15 seconds and 60°C annealing and amplification for 4 min. To remove unincorporated primers, samples were subjected to an ExoI reaction (New England BioLabs). STA-ExoI treated samples were then diluted 1:2 with DNA suspension buffer then mixed with 2X TaqMan Gene expression Mater mix (Applied Biosystems), 20X DNA Binding Dye Sample Loading Reagent (Fluidigm) and 20X EvaGreen DNA binding dye (Biotium) and then loaded onto the dynamic array sample inlet. Assays were prepared by mixing 20 uM of each forward and reverse primer mix (Supplemental Table S2), DNA suspension buffer and 2X Assay Loading Reagent (Fluidigm) and then loaded onto the dynamic array. Using the IFC controller HX (Fluidigm), the samples and assays were loaded and the chip was then placed into the BioMark HD System (Fluidigm) following manufacturer's instructions.
A primary filter of single cell data consisted of determining the correct melting temperature for each primer pair and analyzing the exported Ct values using Fluidigm's SINGuLAR Analysis Toolset 2.0. A limit of detection (LoD) was determined to be Ct value of 26 as described in Fluidigm's Application Guidance: Single-Cell Data Analysis. Log2Ex values are equal to the LoD Ct (26) – Ct of the gene of interest. Log2Ex values then represent the transcript level above background, expressed in log2. ANOVA statistical analysis of single cells was carried out using Fluidigm's SINGuLAR Analysis Toolset 2.0.
RNA Sequencing
Total RNA was extracted using the RNeasy Plus Micro Kit (Qiagen) per manufacturer's instructions. Five ng of total RNA was subjected to first and second strand cDNA synthesis and purification using the Ovation RNA-Seq System V2 (NuGEN Technologies). Next, Single Primer Isolation Amplification (SPIA) and purification was conducted. cDNA was then fragmented using the Covaris S-Series system and Illumina libraries were prepared using the NEBNext DNA Sample Prep Master Mix set. Briefly, fragmented DNA was end repaired, followed by cleanup with Agencourt AMPure XB beads (Beckman Coulter Genomics). End repaired DNA was then dA-tailed followed by adaptor ligation. Samples were sequenced by the Illumina HiSeq 2000 platform (Illumina) as 50 bp single-ended (iPSCs) or 100 bp pair-end reads (H9 hESC and differentiated populations. In order to compare the single- and paired-end runs, only 50 bps of a single end of paired-end sequencing runs were used for mapping. Read information is contained in Supplemental Table S3. The TopHat package54 was used to align the reads to the hg19 reference genome without the Y chromosome. Cufflinks55 was used to calculate RPKM expression values for refSeq annotated genes56 and cummeRbund was used to visualize the data57.
Lentiviral VASA-GFP reporter, transduction and FACS
Undifferentiated iPSCs and hESCs were transduced, selected and differentiated on Matrigel (BD Biosciences) –coated plates with the VASA-GFP reporter (pLVGV) as previously described12,13. Differentiation medium contained 50 ng/ml Bone Morphogenetic Proteins 4 and 8a (BMP4 and BMP8a; R&D Systems) and 10 nM RA and was changed on day 0 and 3. Cells were prepared for FACS as described above. Cells were first sorted for GFP+ populations and then single cells were sorted from the pure population into individual wells of an 8 well strip with 5 ul of Cells Direct 2X reaction mixture (Life Technologies). Bulk GFP+ and GFP- populations were also collected for RNA sequencing analysis.
Xenotransplantation Assay
iPSCs or hESCs were transplanted into the testes of busulfan-treated, immunodeficient nude mice (NCr nu/nu; Tactonic) as previous described19,20,58,59. Briefly, immunodeficient mice were treated with a single dose of busulfan (40 mg/kg, Sigma-Aldrich) at six weeks of age to rid the mouse of endogenous spermatogenesis. Five weeks after treatment, xenotransplantation was performed by injection of 5 to 7 μl of cell suspension containing 10% trypan blue (Invitrogen) into the seminiferous tubules of recipient mouse testes via cannulation of the efferent ducts (7.9 × 105 to 1.6 × 106 cells/recipient mouse testis). Eight weeks post xenotransplantation, the testis were either fixed immediately with 4% PFA or recipient mouse seminiferous tubules were dispersed with Collagenase IV (1 mg/ml; Life Technologies) and DNase I (1 mg/ml; New England Biolabs) in D-PBS, then fixed with 4% PFA.
Immunohistochemistry of recipient mouse testes
PFA fixed mouse testes were sectioned by AML laboratories (Baltimore, MD) by paraffin embedding and conducting serial cross-sectioning every 5 μm. Deparafinization was conducted by two consecutive 10 min. xylenes (Sigma-Aldrich) treatments followed by rehydration in 100%, 100%, 90%, 80% and 70% ethanol treatments followed by a 10 minute wash in tap water. Antigen retrieval was conducted by boiling slides for half hour in 0.01 M Sodium Citrate (pH 6.0; Sigma-Aldrich), then cooling slides for an additional half hour, followed by a 10 min. was in PBS. Blocking and permeabilization was conducted by addition of 10% normal Donkey serum (Jackson Immunoreearch) with 0.1% Triton X (Sigma-Aldrich) in PBS for 1 hour, followed by incubation with the following primary antibodies diluted in 1% blocking solution overnight at 4°C in a humidified chamber: NUMA (Abcam), cKIT (Dako), H3K27me3 (Millipore), VASA (R&D), OCT4 (Santa Cruz), STELLA (Abcam) and DAZL (Novus). Slides were washed with PBST, followed by an hour incubation with fluorescently labeled secondary antibodies raised in Donkey, followed by additional washes with PBST. All samples were mounted with ProLong Gold Anti-fade mounting media containing DAPI (Life Technologies). Samples were then imaged using a confocal microscope (Zeiss). Quantification of NUMA/VASA double positive staining was determined manually from multiple sections taken from 2–3 different depths within the testis (technical replicates; Supplemental Fig. S7C and S7D) and from multiple clonal replicates for each transplanted cell line (biological replicates; Fig. 5C and 5D). Sample numbers were as follows: H9 – 4, Triple X – 6, Control subclone 8 – 4 and TSC1 subclone 2 – 10. Counts were only taken from whole testis (Supplemental Table S4) and if the testis contained NUMA+/VASA+ cells – i.e. zeros were not factored into total counts (Supplemental Fig. S7B and Supplemental Table S4).
Additional Information
Accession codes: All RNA-Seq reads are available at the GEO database with accession number GSE55939.
References
Hassold, T. et al. Human aneuploidy: incidence, origin and etiology. Environ Mol Mutagen 28, 167–175 (1996).
Davenport, M. L. et al. Growth failure in early life: an important manifestation of Turner syndrome. Horm Res 57 157–164 (2002).
Bondy, C. A. Heart disease in Turner syndrome. Minerva Endocrinol 32, 245–261 (2007).
Ford, C. E., Jones, K. W., Polani, P. E., De Almeida, J. C. & Briggs, J. H. A sex-chromosome anomaly in a case of gonadal dysgenesis (Turner's syndrome). Lancet 1, 711–713 (1959).
Turner, H. H. A Syndrome of Infantilism, Congenital Webbed Neck and Cubitus Valgus. Endocrinology 23, 566–574 (1938).
Dewhurst, J. Fertility in 47,XXX and 45,X patients. J Med Genet 15, 132–135 (1978).
Carrel, L. & Willard, H. F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005).
Rao, E., Weiss, B., Fukami, M., Rump, A., Niesler, B., Mertz, A., Muroya, K., Binder, G., Kirsch, S. & Winkelmann, M. et al. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet 16, 54–63 (1997).
Lyon, M. F. & Hawker, S. G. Reproductive lifespan in irradiated and unirradiated chromosomally XO mice. Genet Res 21, 185–194 (1973).
Burgoyne, P. S. & Baker, T. G. Oocyte depletion in XO mice and their XX sibs from 12 to 200 days post partum. J Reprod Infertil 61, 207–212 (1981).
Burgoyne, P. S. & Baker, T. G. Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. J Reprod Infertil 75, 633–645 (1985).
Speed, R. M. Oocyte development in XO foetuses of man and mouse: the possible role of heterologous X-chromosome pairing in germ cell survival. Chromosoma 94, 115–124 (1986).
Adler, D. A., Bressler, S. L., Chapman, V. M., Page, D. C. & Disteche, C. M. Inactivation of the Zfx gene on the mouse X chromosome. PNAS 88, 4592–4595 (1991).
Ashworth, A., Rastan, S., Lovell-Badge, R. & Kay, G. X-chromosome inactivation may explain the difference in viability of XO humans and mice. Nature 351, 406–408 a0 (1991).
Panula, S. et al. Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum Mol Genet 20, 752–762 (2011).
Kee, K., Angeles, V. T., Flores, M., Nguyen, H. N. & Reijo Pera, R. A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 462, 222–225 (2009).
Medrano, J. V., Ramathal, C., Nguyen, H. N., Simon, C. & Reijo Pera, R. A. Divergent RNA-binding proteins, DAZL and VASA, induce meiotic progression in human germ cells derived in vitro. Stem Cells 30, 441–451 (2012).
Gkountela, S. et al. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol 15, 113–122 (2013).
Durruthy Durruthy, J. et al. Fate of induced pluripotent stem cells following transplantation to murine seminiferous tubules. Hum Mol Genet 23, 3071–3084 (2014).
Ramathal, C. et al. Fate of iPSCs Derived from Azoospermic and Fertile Men following Xenotransplantation to Murine Seminiferous Tubules. Cell Rep 7, 1284–1297 (2014).
Magnusdottir, E. et al. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat Cell Biol 15, 905–915 (2013).
Nakaki, F. et al. Induction of mouse germ-cell fate by transcription factors in vitro. Nature 501, 222–226 (2013).
Hayashi, K., Ohta, H., Kurimoto, K., Aramaki, S. & Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532 (2011).
Hayashi, K. et al. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science 338, 971–975 (2012).
Nicholas, C. R., Haston, K. M., Grewall, A. K., Longacre, T. A. & Reijo Pera, R. A. Transplantation directs oocyte maturation from embryonic stem cells and provides a therapeutic strategy for female infertility. Hum Mol Genet 18, 4376–4389 (2009).
Brinster, R. L. & Avarbock, M. R. Germline transmission of donor haplotype following spermatogonial transplantation. PNAS 91, 11303–11307 (1994).
Brinster, R. L. Germline stem cell transplantation and transgenesis. Science 296, 2174–2176 (2002).
Nagano, M., Patrizio, P. & Brinster, R. L. Long-term survival of human spermatogonial stem cells in mouse testes. Fertil Steril 78, 1225–1233 (2002).
Ogawa, T. Spermatogonial transplantation: the principle and possible applications. J Mol Med 79, 368–374 (2001).
Somers, A. et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem cells 28, 1728–1740 (2010).
Huangfu, D. et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 26, 1269–1275 (2008).
Henderson, J. K. et al. Preimplantation human embryos and embryonic stem cells show comparable expression of stage-specific embryonic antigens. Stem Cells 20, 329–337 (2002).
Li, W. et al. Modeling abnormal early development with induced pluripotent stem cells from aneuploid syndromes. Hum Mol Genet 21, 32–45 (2012).
Ross, M. T. et al. The DNA sequence of the human X chromosome. Nature 434, 325–337 (2005).
Brown, C. J. et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542 (1992).
Brown, C. J. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44 (1991).
Plath, K. et al. Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 (2003).
Lessing, D., Anguera, M. C. & Lee, J. T. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet 14, 85–110 (2013).
Mekhoubad, S. et al. Erosion of dosage compensation impacts human iPSC diseasemodeling. Cell stem cell 10, 595–609 (2012).
Kim, K. Y. et al. X Chromosome of Female Cells Shows Dynamic Changes in Status during Human Somatic Cell Reprogramming. Stem cell reports 2, 896–909 (2014).
Lawson, K. A. et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev 13, 424–36 (1999).
Hiller, M., Liu, C., Blumenthal, P. D., Gearhart, J. L. & Kerr, C. L. Bone morphogenetic protein 4 mediates human embryonic germ cell derivation. Stem Cells Dev. 20, 351–361 (2011).
Hayashi, Y., Saitou, M. & Yamanaka, S. Germline development from human pluripotent stem cells toward disease modeling of infertility. Fertil Steril 97, 1250–1259 (2012).
Saadai, P. et al. Human induced pluripotent stem cell-derived neural crest stem cells integrate into the injured spinal cord in the fetal lamb model of myelomeningocele. J Pediatr Surg 48, 158–163 (2013).
Lydersen, B. K. & Pettijohn, D. E. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell 22, 489–499 (1980).
Gkountela, S. et al. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol 15, 113–122 (2013).
Anderson, R. A., Fulton, N., Cowan, G., Coutts, S. & Saunders, P. T. Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev Biol 7, 136 (2007).
Wongtrakoongate, P., Jones, M., Gokhale, P. J. & Andrews, P. W. STELLA facilitates differentiation of germ cell and endodermal lineages of human embryonic stem cells. PloS one 8, e56893 (2013).
McLaren, A. Meiosis and differentiation of mouse germ cells. Symp Soc Exp Biol 38, 7–23 (1984).
Adams, I. R. & McLaren, A. Sexually dimorphic development of mouse primordial germ cells: switching from oogenesis to spermatogenesis. Development 129, 1155–1164 (2002).
Palmer, S. J. & Burgoyne, P. S. XY follicle cells in the ovaries of XO/XY and XO/XY/XYY mosaic mice. Development 111, 1017–1019 (1991).
Byrne, J. A., Nguyen, H. N. & Reijo Pera, R. A. Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. PloS one 4, e7118 (2009).
Nguyen, H. N. et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell stem cell 8, 267–280 (2011).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010).
Pruitt, K. D., Tatusova, T., Brown, G. R. & Maglott, D. R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–135 (2012).
Goff, L., Trapnell, C. & Kelley, D. R. CummeRbund: Visualization and Exploration of Cufflinks High-throughput Sequencing Data. R Package Version 2.2.0 (2012).
Hermann, B. P. et al. Characterization, cryopreservation and ablation of spermatogonial stem cells in adult rhesus macaques. Stem Cells 25, 2330–2338 (2007).
Dovey, S. L. et al. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J Clin Invest 123, 1833–1843 (2013).
Acknowledgements
We thank members of the Reijo Pera laboratory for guidance and discussions. In particular, we are grateful to C. Ramathal, J. Cui and J. Durruthy Durruthy for assistance and guidance during xenotransplantation analysis and for male xenotransplant data. We thank S. Chavez and S. Schuh-Huerta for discussions throughout the project, M. Moncada for assistance with iPSC characterization, D. Yang for assistance with RNA-Seq sample preparation and S. Briggs for assistance with RNA FISH. We are thankful to M. Angueara and J. Lee for the Triple X iPSC line. This study was supported by the National Institute of Child Health and Human Development (NICHD U54 HD068158) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research. H. R. Chiang is supported by Stanford CIRM Training Program (TG2-01159).
Author information
Authors and Affiliations
Contributions
A.A.D. and R.A.R.P. conceived and designed experiments. A.A.D. performed experiments. H.R.C analyzed RNAseq data. M.S. and K.E.O. performed xenotransplantation experiments. A.A.D. and R.A.R.P. wrote the paper. All authors edited the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Dominguez, A., Chiang, H., Sukhwani, M. et al. Human germ cell formation in xenotransplants of induced pluripotent stem cells carrying X chromosome aneuploidies. Sci Rep 4, 6432 (2014). https://doi.org/10.1038/srep06432
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06432
This article is cited by
-
Competing endogenous RNA network analysis of Turner syndrome patient-specific iPSC-derived cardiomyocytes reveals dysregulation of autosomal heart development genes by altered dosages of X-inactivation escaping non-coding RNAs
Stem Cell Research & Therapy (2023)
-
A PAX5–OCT4–PRDM1 developmental switch specifies human primordial germ cells
Nature Cell Biology (2018)
-
Uniparental disomy of the entire X chromosome in Turner syndrome patient-specific induced pluripotent stem cells
Cell Discovery (2015)
-
Dynamic and social behaviors of human pluripotent stem cells
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.